For calculations related to the loaded allele effects and internal additive breeding value model. More...
Functions | |
| gsc_GroupNum | gsc_split_by_bv (gsc_SimData *d, const gsc_GroupNum group, const gsc_EffectID effID, const unsigned int top_n, const _Bool lowIsBest) |
Takes the top_n individuals in the group with the best breeding values/fitnesses and puts them in a new group. More... | |
| gsc_DecimalMatrix | gsc_calculate_bvs (const gsc_SimData *d, const gsc_GroupNum group, const gsc_EffectID effID) |
| Calculate the fitness metric/breeding value for every genotype in the simulation or every genotype in a certain group. More... | |
| gsc_DecimalMatrix | gsc_calculate_utility_bvs (gsc_BidirectionalIterator *targets, const gsc_EffectMatrix *effset) |
| Calculate the fitness metric/breeding value for a set of genotypes. More... | |
| gsc_DecimalMatrix | gsc_calculate_allele_counts (const gsc_SimData *d, const gsc_GroupNum group, const char allele) |
| Calculates the number of times at each marker that a particular allele appears. More... | |
| void | gsc_calculate_utility_allele_counts (const unsigned int n_markers, const unsigned int n_genotypes, const char **const genotypes, const char allele, gsc_DecimalMatrix *counts) |
| Calculates the number of times at each marker that a particular allele appears. More... | |
| void | gsc_calculate_utility_allele_counts_pair (const unsigned int n_markers, const unsigned int n_genotypes, const char **const genotypes, const char allele, gsc_DecimalMatrix *counts, const char allele2, gsc_DecimalMatrix *counts2) |
| Calculates the number of times at each marker that two particular alleles appear. More... | |
| gsc_MarkerBlocks | gsc_create_evenlength_blocks_each_chr (const gsc_SimData *d, const gsc_MapID mapid, const unsigned int n) |
| Divide the genotype into blocks where each block contains all markers within a 1/n length section of each chromosome in the map, and return the resulting blocks in a struct. More... | |
| gsc_MarkerBlocks | gsc_load_blocks (const gsc_SimData *d, const char *block_file) |
| Given a file containing definitions of blocks of markers, process that file and return a struct containing the definitions of those blocks. More... | |
| void | gsc_calculate_group_local_bvs (const gsc_SimData *d, const gsc_MarkerBlocks b, const gsc_EffectID effID, const char *output_file, const gsc_GroupNum group) |
| Given a set of blocks of markers in a file, for each genotype in a group, calculate the local fitness metric/breeding value for the first allele at each marker in the block, and the local fitness metric/breeding value for the second allele at each marker in the block, then save the result to a file. More... | |
| void | gsc_calculate_local_bvs (const gsc_SimData *d, const gsc_MarkerBlocks b, const gsc_EffectID effID, const char *output_file) |
| Given a set of blocks of markers in a file, for each genotype saved, calculate the local BV for the first allele at each marker in the block, and the local BV for the second allele at each marker in the block, then save the result to a file. More... | |
| char * | gsc_calculate_optimal_haplotype (const gsc_SimData *d, const gsc_EffectID effID) |
| Takes a look at the currently-loaded effect values and creates a string containing the allele with the highest effect value for each marker as ordered in the gsc_SimData. More... | |
| char * | gsc_calculate_optimal_possible_haplotype (const gsc_SimData *d, const gsc_GroupNum group, const gsc_EffectID effID) |
| Calculates the highest-breeding-value haplotype that can be created from the alleles present in a given group. More... | |
| double | gsc_calculate_optimal_bv (const gsc_SimData *d, const gsc_EffectID effID) |
| Takes a look at the currently-loaded effect values and returns the highest possible breeding value any (diploid) genotype could have using those effect values. More... | |
| double | gsc_calculate_optimal_possible_bv (const gsc_SimData *d, const gsc_GroupNum group, const gsc_EffectID effID) |
| Calculates the breeding value of the highest breeding-value genotype that can be created from the alleles present in a given group. More... | |
| double | gsc_calculate_minimal_bv (const gsc_SimData *d, const gsc_EffectID effID) |
| Takes a look at the currently-loaded effect values and returns the lowest possible breeding value any (diploid) genotype could score using those effect values. More... | |
Detailed Description
For calculations related to the loaded allele effects and internal additive breeding value model.
Function Documentation
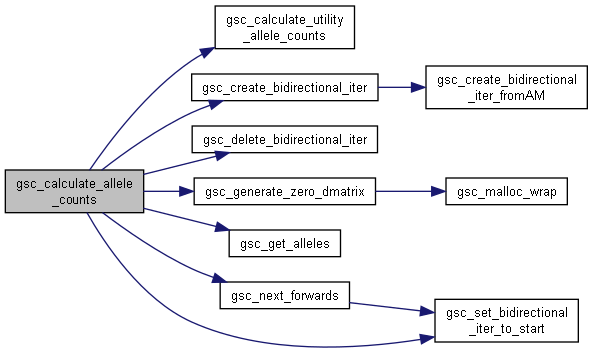
◆ gsc_calculate_allele_counts()
| gsc_DecimalMatrix gsc_calculate_allele_counts | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const char | allele | ||
| ) |
Calculates the number of times at each marker that a particular allele appears.
Returns the results in a calculated DecimalMatrix. This structure should be deleted with delete_dmatrix once there is no more use for it.
Has short name: calculate_allele_counts
- Parameters
-
d simulation object containing the genotypes whose alleles will be counted group NO_GROUP to count alleles of all genotypes in the simulation, or a specific group identifier to count only the alleles of members of that group. allele the single-character allele to be counting.
- Returns
- a (num genotypes x num markers) gsc_DecimalMatrix, with each cell containing the number of incidences of the allele. The first axis of the DecimalMatrix corresponds to candidate genotypes, the second axis corresponds to genetic markers.
Definition at line 9188 of file sim-operations.c.
 Here is the call graph for this function:
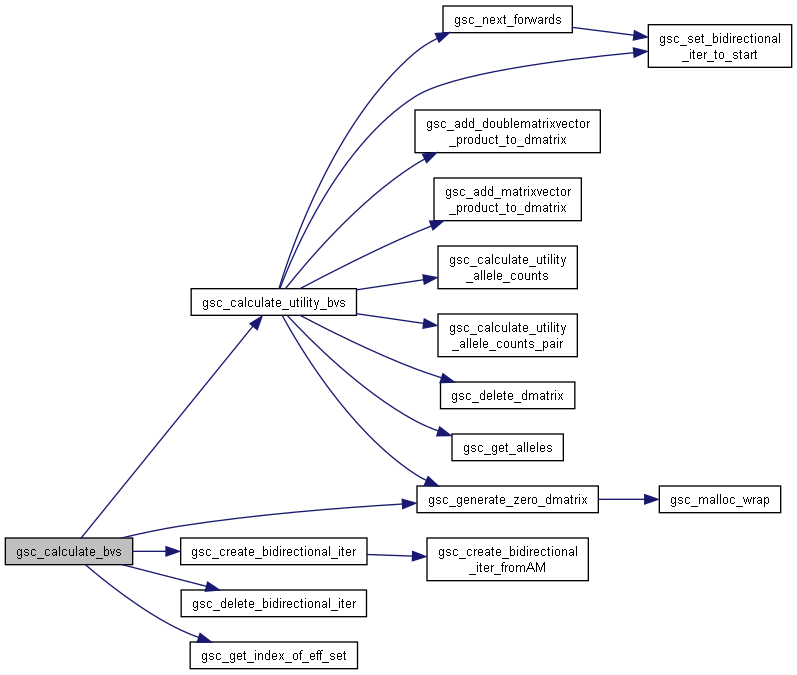
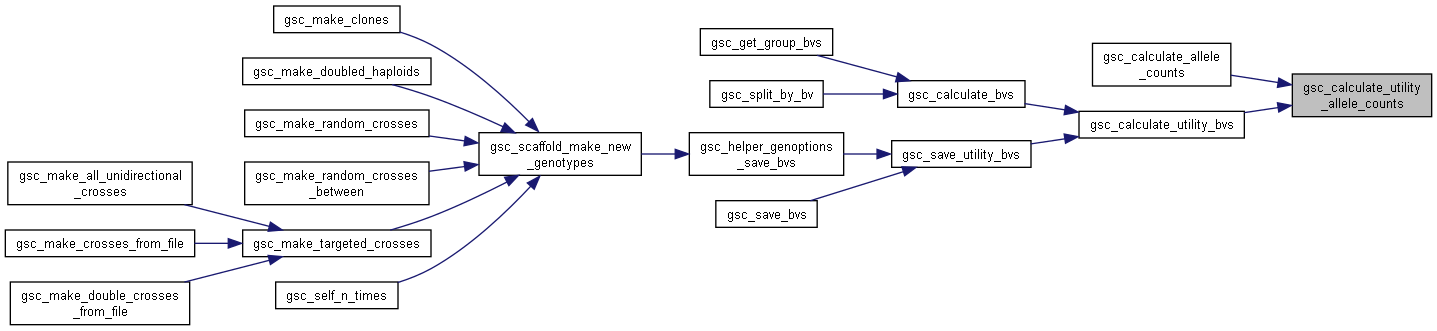
Here is the call graph for this function:◆ gsc_calculate_bvs()
| gsc_DecimalMatrix gsc_calculate_bvs | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_EffectID | effID | ||
| ) |
Calculate the fitness metric/breeding value for every genotype in the simulation or every genotype in a certain group.
To calculate the breeding value, the number of copies of each allele at each marker for each genotype are counted. The counts of each allele are multiplied by the additive marker effect of that allele at that marker. Then, each genotype's allele effects across all markers are summed up.
Has short name: calculate_bvs
- Parameters
-
d pointer to the gsc_SimData object in which the marker effects and genotypes to be used to calculate breeding values are stored. group NO_GROUP to calculate breeding values for all genotypes in the simulation, or a group number to calculate breeding values for all members of that group. effID Identifier of the marker effect set to be used to calculate these breeding values
- Returns
- A (1 x number of genotypes) gsc_DecimalMatrix containing the breeding value scores
Definition at line 9093 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_calculate_group_local_bvs()
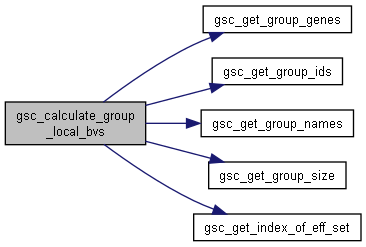
| void gsc_calculate_group_local_bvs | ( | const gsc_SimData * | d, |
| const gsc_MarkerBlocks | b, | ||
| const gsc_EffectID | effID, | ||
| const char * | output_file, | ||
| const gsc_GroupNum | group | ||
| ) |
Given a set of blocks of markers in a file, for each genotype in a group, calculate the local fitness metric/breeding value for the first allele at each marker in the block, and the local fitness metric/breeding value for the second allele at each marker in the block, then save the result to a file.
This gives block effects/local BVs for each haplotype of each individual in the group.
The output file will have format:
[genotype name]_1 [effect for first block, first allele] [effect for second block, second allele] ...
[genotype name]_2 [effect for first block, second allele] [effect for second block, second allele] ...
[genotype 2 name]_1 ...
Has short name: calculate_group_local_bvs
- Parameters
-
d pointer to the gsc_SimData object to which the groups and individuals belong. It must have a marker effect file loaded to successfully run this function. b struct containing the blocks to use effID Identifier of the marker effect set to be used to calculate these breeding values output_file string containing the filename of the file to which output block effects/local BVs will be saved. group group number from which to split the top individuals.
Definition at line 9593 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_local_bvs()
| void gsc_calculate_local_bvs | ( | const gsc_SimData * | d, |
| const gsc_MarkerBlocks | b, | ||
| const gsc_EffectID | effID, | ||
| const char * | output_file | ||
| ) |
Given a set of blocks of markers in a file, for each genotype saved, calculate the local BV for the first allele at each marker in the block, and the local BV for the second allele at each marker in the block, then save the result to a file.
This gives block effects for each haplotype of each individual currently saved to the gsc_SimData.
The output file will have format:
[genotype name]_1 [effect for first block, first allele] [effect for second block, second allele] ...
[genotype name]_2 [effect for first block, second allele] [effect for second block, second allele] ...
[genotype 2 name]_1 ...
Has short name: calculate_local_bvs
- Parameters
-
d pointer to the gsc_SimData object to which individuals belong. It must have a marker effect file loaded to successfully run this function. b struct containing the blocks to use effID Identifier of the marker effect set to be used to calculate these breeding values output_file string containing the filename of the file to which output block effects/local BV will be saved.
Definition at line 9710 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_minimal_bv()
| double gsc_calculate_minimal_bv | ( | const gsc_SimData * | d, |
| const gsc_EffectID | effID | ||
| ) |
Takes a look at the currently-loaded effect values and returns the lowest possible breeding value any (diploid) genotype could score using those effect values.
The gsc_SimData must be initialised with marker effects for this function to succeed.
Has short name: calculate_minimal_bv
- Parameters
-
d pointer to the gsc_SimData containing markers and marker effects. effID Identifier of the marker effect set to be used to calculate these breeding values
- Returns
- the fitness metric/breeding value of the worst genotype.
Definition at line 10055 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_optimal_bv()
| double gsc_calculate_optimal_bv | ( | const gsc_SimData * | d, |
| const gsc_EffectID | effID | ||
| ) |
Takes a look at the currently-loaded effect values and returns the highest possible breeding value any (diploid) genotype could have using those effect values.
The gsc_SimData must be initialised with marker effects for this function to succeed.
Has short name: calculate_optimal_bv
- Parameters
-
d pointer to the gsc_SimData containing markers and marker effects. effID Identifier of the marker effect set to be used to calculate these breeding values
- Returns
- the fitness metric/breeding value of the best/ideal genotype.
Definition at line 9931 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_optimal_haplotype()
| char * gsc_calculate_optimal_haplotype | ( | const gsc_SimData * | d, |
| const gsc_EffectID | effID | ||
| ) |
Takes a look at the currently-loaded effect values and creates a string containing the allele with the highest effect value for each marker as ordered in the gsc_SimData.
Returns this as a null-terminated heap array of characters of length d->n_markers + one null byte.
The return value should be freed when usage is finished!
The gsc_SimData must be initialised with marker effects for this function to succeed.
Has short name: calculate_optimal_haplotype
- Parameters
-
d pointer to the gsc_SimData containing markers and marker effects. effID Identifier of the marker effect set to be used to calculate these breeding values
- Returns
- a heap array filled with a null-terminated string containing the highest scoring allele at each marker.
Definition at line 9809 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_optimal_possible_bv()
| double gsc_calculate_optimal_possible_bv | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_EffectID | effID | ||
| ) |
Calculates the breeding value of the highest breeding-value genotype that can be created from the alleles present in a given group.
The highest value genotype is completely homozygous with the same alleles as the haplotype from gsc_calculate_optimal_possible_haplotype(), because of the additive model of trait effects.
The gsc_SimData must be initialised with marker effects for this function to succeed.
Has short name: calculate_optimal_possible_bv
- Parameters
-
d pointer to the gsc_SimData containing markers and marker effects. group number to look at the available alleles in effID Identifier of the marker effect set to be used to calculate these breeding values
- Returns
- the fitness metric/breeding value of the best genotype
Definition at line 9973 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_optimal_possible_haplotype()
| char * gsc_calculate_optimal_possible_haplotype | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_EffectID | effID | ||
| ) |
Calculates the highest-breeding-value haplotype that can be created from the alleles present in a given group.
The return value should be freed when usage is finished!
The gsc_SimData must be initialised with marker effects for this function to succeed.
Has short name: calculate_optimal_possible_haplotype
- Parameters
-
d pointer to the gsc_SimData containing markers and marker effects. group group number to look at the available alleles in effID Identifier of the marker effect set to be used to calculate these breeding values
- Returns
- a heap array filled with a null-terminated string containing the highest scoring allele at each marker.
Definition at line 9850 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_calculate_utility_allele_counts()
| void gsc_calculate_utility_allele_counts | ( | const unsigned int | n_markers, |
| const unsigned int | n_genotypes, | ||
| const char **const | genotypes, | ||
| const char | allele, | ||
| gsc_DecimalMatrix * | counts | ||
| ) |
Calculates the number of times at each marker that a particular allele appears.
Saves the result to a gsc_DecimalMatrix provided by the calling context.
- Parameters
-
n_markers number of genetic markers in each genotype. n_genotypes number of genotypes to calculate counts for. (length of the following vector) genotypes pointers to genotype strings for which to count the alleles at particular markers. allele the single-character allele to be counting. counts pointer to the gsc_DecimalMatrix into which to put the number of alleleoccurences at each row/marker for each column/genotype in the group. The DecimalMatrix should have at least n_genotypes rows and n_markers columns, if not, no output will be saved.
Definition at line 9232 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_calculate_utility_allele_counts_pair()
| void gsc_calculate_utility_allele_counts_pair | ( | const unsigned int | n_markers, |
| const unsigned int | n_genotypes, | ||
| const char **const | genotypes, | ||
| const char | allele, | ||
| gsc_DecimalMatrix * | counts, | ||
| const char | allele2, | ||
| gsc_DecimalMatrix * | counts2 | ||
| ) |
Calculates the number of times at each marker that two particular alleles appear.
Saves the results for each allele to a gsc_DecimalMatrix provided by the calling context.
This function is the same as gsc_calculate_utility_allele_counts, but for two alleles at a time, for the purpose of loop unrolling in breeding value calculations.
- Parameters
-
n_markers number of genetic markers in each genotype. n_genotypes number of genotypes to calculate counts for. (length of the following vector) genotypes pointers to genotype strings for which to count the alleles at particular markers. allele the first single-character allele to be counting. counts pointer to the gsc_DecimalMatrix into which to put the number of alleleoccurences at each row/marker for each column/genotype in the group. The DecimalMatrix should have at least n_genotypes rows and n_markers columns, if not, no output will be saved.allele2 the second single-character allele to be counting. counts2 pointer to the gsc_DecimalMatrix into which to put the number of allele2occurences at each row/marker for each column/genotype in the group. The DecimalMatrix should have at least n_genotypes rows and n_markers columns, if not, no output will be saved.
Definition at line 9280 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_calculate_utility_bvs()
| gsc_DecimalMatrix gsc_calculate_utility_bvs | ( | gsc_BidirectionalIterator * | targets, |
| const gsc_EffectMatrix * | effset | ||
| ) |
Calculate the fitness metric/breeding value for a set of genotypes.
The breeding value is calculated as the sum, across all alleles, of the (genotypes x markers) count matrix for that allele multiplied by the (markers x 1) marker effects for that allele.
- Parameters
-
targets Iterator for the genotypes whose breeding values are to be calculated. effset Set of additive marker effects to use to calculate the breeding values.
- Returns
- (1 x number of genotypes in iterator) gsc_DecimalMatrix, containing the breeding value scores
Definition at line 9123 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_create_evenlength_blocks_each_chr()
| gsc_MarkerBlocks gsc_create_evenlength_blocks_each_chr | ( | const gsc_SimData * | d, |
| const gsc_MapID | mapid, | ||
| const unsigned int | n | ||
| ) |
Divide the genotype into blocks where each block contains all markers within a 1/n length section of each chromosome in the map, and return the resulting blocks in a struct.
Chromosomes where there are no markers tracked are excluded/no blocks are created in those chromosomes.
Chromosomes where only one marker is tracked put the marker in the first block, and have all other blocks empty.
Empty blocks have blocks.num_markers_in_block[index] equal to 0 and contain a null pointer at blocks.markers_in_block[index].
Remember to call the gsc_MarkerBlocks destructor gsc_delete_markerblocks() on the returned struct.
Has short name: create_evenlength_blocks_each_chr
- Parameters
-
d pointer to the gsc_SimData object to which the groups and individuals belong. It must have a marker effect file loaded to successfully run this function. mapid ID of the recombination map whose markers and chromosomes will be divided into blocks, or NO_MAP to use the first-loaded/primary map by default. mapid recombination map to use to determine block lengths and marker allocations to blocks. If this value is NO_MAP, uses first/primary map. n number of blocks into which to split each chromosome.
- Returns
- a struct containing the markers identified as belonging to each block.
Definition at line 9340 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_load_blocks()
| gsc_MarkerBlocks gsc_load_blocks | ( | const gsc_SimData * | d, |
| const char * | block_file | ||
| ) |
Given a file containing definitions of blocks of markers, process that file and return a struct containing the definitions of those blocks.
The block file is designed after the output from a call to the R SelectionTools package's st.def.hblocks function. It should have the format (tab-separated):
Chrom Pos Name Class Markers
[ignored] [ignored] [ignored] [ignored] [semicolon];[separated];[list] ;[of];[marker];[names];[belonging];[to];[this];[block];
...
Has short name: load_blocks
- Parameters
-
d pointer to the gsc_SimData object to which the groups and individuals belong. It must have a marker effect file loaded to successfully run this function. block_file string containing filename of the file with blocks
- Returns
- a struct containing the markers identified as belonging to each block according to their definitions in the file.
Definition at line 9495 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_by_bv()
| gsc_GroupNum gsc_split_by_bv | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_EffectID | effID, | ||
| const unsigned int | top_n, | ||
| const _Bool | lowIsBest | ||
| ) |
Takes the top_n individuals in the group with the best breeding values/fitnesses and puts them in a new group.
The new group number is returned.
Has short name: split_by_bv
- Parameters
-
d pointer to the gsc_SimData object to which the groups and individuals belong. It must have a marker effect file loaded to successfully run this function. group group number from which to split the top individuals. effID Identifier of the set of marker effects to use to calculate BV. top_n The number of individuals to put in the new group. lowIsBest boolean, if TRUE the top_nwith the lowest breeding value will be selected, if false thetop_nwith the highest breeding value are.
- Returns
- the group number of the newly-created split-off group
Definition at line 9020 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: