Composite type that is used to run crossing simulations. More...
#include <sim-operations.h>
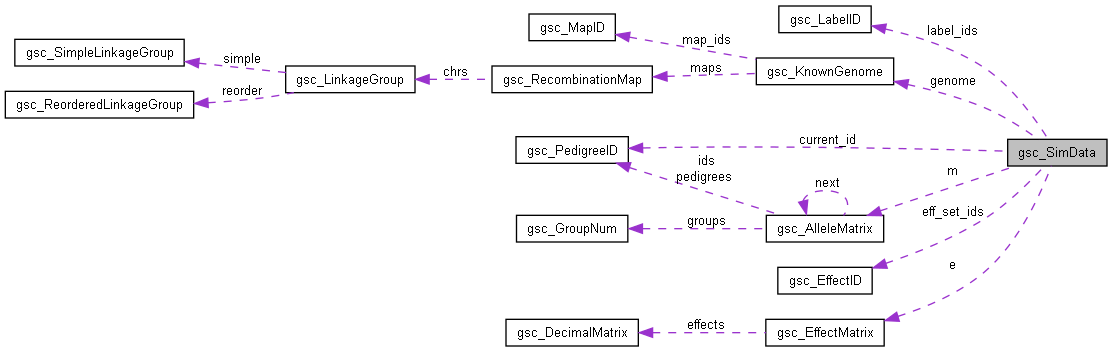
 Collaboration diagram for gsc_SimData:
Collaboration diagram for gsc_SimData:Data Fields | |
| unsigned int | n_labels |
| The number of custom labels in the simulation. More... | |
| gsc_LabelID * | label_ids |
| The identifier number of each label in the simulation, in order of their lookup index. More... | |
| int * | label_defaults |
| Array containing the default (birth) value of each custom label. More... | |
| gsc_KnownGenome | genome |
| A gsc_KnownGenome, which stores the information of known markers and linkage groups, as well as one or more recombination maps for use in simulating meiosis. More... | |
| gsc_AlleleMatrix * | m |
| Pointer to an gsc_AlleleMatrix, which stores data and metadata of founders and simulated offspring. More... | |
| unsigned int | n_eff_sets |
| The number of sets of allele effects in the simulation. More... | |
| gsc_EffectID * | eff_set_ids |
| The identifier number of each set of allele effects in the simulation, ordered by their lookup index. More... | |
| gsc_EffectMatrix * | e |
| Array of n_eff_sets gsc_EffectMatrix, optional for the use of the simulation. More... | |
| rnd_pcg_t | rng |
| Random number generator working memory. More... | |
| gsc_PedigreeID | current_id |
| Highest SimData-unique ID that has been generated so far. More... | |
| unsigned int | n_groups |
| Number of groups currently existing in simulation. More... | |
Detailed Description
Composite type that is used to run crossing simulations.
The core of this type is a list of markers. These are used to index the rows of the allele matrix and the position map, and the columns of the effect matrix.
Has short name: SimData
Definition at line 834 of file sim-operations.h.
Field Documentation
◆ current_id
| gsc_PedigreeID gsc_SimData::current_id |
Highest SimData-unique ID that has been generated so far.
Used to track which IDs have already been given out.
Definition at line 857 of file sim-operations.h.
◆ e
| gsc_EffectMatrix* gsc_SimData::e |
Array of n_eff_sets gsc_EffectMatrix, optional for the use of the simulation.
Used for calculating breeding values from which alleles a genotype has at each marker.
Definition at line 852 of file sim-operations.h.
◆ eff_set_ids
| gsc_EffectID* gsc_SimData::eff_set_ids |
The identifier number of each set of allele effects in the simulation, ordered by their lookup index.
Definition at line 850 of file sim-operations.h.
◆ genome
| gsc_KnownGenome gsc_SimData::genome |
A gsc_KnownGenome, which stores the information of known markers and linkage groups, as well as one or more recombination maps for use in simulating meiosis.
Definition at line 841 of file sim-operations.h.
◆ label_defaults
| int* gsc_SimData::label_defaults |
Array containing the default (birth) value of each custom label.
Definition at line 838 of file sim-operations.h.
◆ label_ids
| gsc_LabelID* gsc_SimData::label_ids |
The identifier number of each label in the simulation, in order of their lookup index.
Definition at line 836 of file sim-operations.h.
◆ m
| gsc_AlleleMatrix* gsc_SimData::m |
Pointer to an gsc_AlleleMatrix, which stores data and metadata of founders and simulated offspring.
The gsc_AlleleMatrix is start of a linked list if there are many genotypes.
Definition at line 844 of file sim-operations.h.
◆ n_eff_sets
| unsigned int gsc_SimData::n_eff_sets |
The number of sets of allele effects in the simulation.
Definition at line 849 of file sim-operations.h.
◆ n_groups
| unsigned int gsc_SimData::n_groups |
Number of groups currently existing in simulation.
It is guaranteed to never be less than the number of groups in simulation even if not perfectly accurate.
Definition at line 860 of file sim-operations.h.
◆ n_labels
| unsigned int gsc_SimData::n_labels |
The number of custom labels in the simulation.
Definition at line 835 of file sim-operations.h.
◆ rng
| rnd_pcg_t gsc_SimData::rng |
Random number generator working memory.
Definition at line 856 of file sim-operations.h.
The documentation for this struct was generated from the following file: