These functions return vector data, rather than data on a single genotype or single group. More...
 Collaboration diagram for Collective Data Access Functions:
Collaboration diagram for Collective Data Access Functions:Functions | |
| unsigned int | gsc_get_group_size (const gsc_SimData *d, const gsc_GroupNum group_id) |
| Function to count the number of genotypes that currently belong to the specified group. More... | |
| unsigned int | gsc_get_group_genes (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, char **output) |
| Gets a shallow copy of the genes/alleles of each member of the group. More... | |
| unsigned int | gsc_get_group_names (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, char **output) |
| Gets a shallow copy of the names of each member of the group. More... | |
| unsigned int | gsc_get_group_ids (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, gsc_PedigreeID *output) |
| Gets the ids of each member of the group. More... | |
| unsigned int | gsc_get_group_indexes (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, unsigned int *output) |
| Gets the 0-based global indexes of each member of the group. More... | |
| unsigned int | gsc_get_group_bvs (const gsc_SimData *d, const gsc_GroupNum group_id, const gsc_EffectID effID, unsigned int group_size, double *output) |
| Gets the breeding values/breeding values/fitnesses of each member of the group. More... | |
| unsigned int | gsc_get_group_parent_ids (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, const int whichParent, gsc_PedigreeID *output) |
| Gets the ids of either the first or second parent of each member of the group. More... | |
| unsigned int | gsc_get_group_parent_names (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, const int whichParent, char **output) |
| Gets the names of either the first or second parent of each member of the group. More... | |
| unsigned int | gsc_get_group_pedigrees (const gsc_SimData *d, const gsc_GroupNum group_id, unsigned int group_size, char **output) |
| Gets the full pedigree string (as per gsc_save_group_full_pedigree() ) of each member of the group. More... | |
| size_t | gsc_get_existing_groups (gsc_SimData *d, gsc_GroupNum *output) |
| Identify group numbers that currently have members. More... | |
| size_t | gsc_get_existing_group_counts (gsc_SimData *d, gsc_GroupNum *out_groups, unsigned int *out_sizes) |
| Identify group numbers that currently have members, and how many members they have. More... | |
Detailed Description
These functions return vector data, rather than data on a single genotype or single group.
The get_group_ family are left here for legacy purposes: using an iterator is the new and less memory intensive way to do the tasks these were used for.
Function Documentation
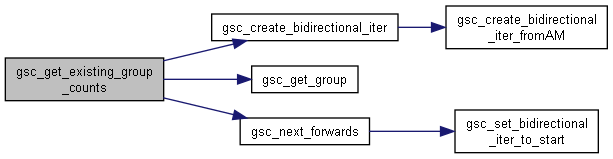
◆ gsc_get_existing_group_counts()
| size_t gsc_get_existing_group_counts | ( | gsc_SimData * | d, |
| gsc_GroupNum * | out_groups, | ||
| unsigned int * | out_sizes | ||
| ) |
Identify group numbers that currently have members, and how many members they have.
It saves the extant groups to the output vectors, and also realigns d->n_groups with the true number of groups.
Has short name: get_existing_group_counts
- Parameters
-
d the gsc_SimData struct on which to perform the operation out_groups Pointer to a location to save the vector of extant groups. It must have at least space for d->n_groups group numbers. Else, if it is NULL, then the group numbers of existing groups are not saved. out_sizes Pointer to a location to save the vector of those groups' sizes. It must have at least space for d->n_groups group sizes. Else, if it is NULL, then the counts of existing groups are not saved.
- Returns
- The number of existing groups.
Definition at line 3559 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
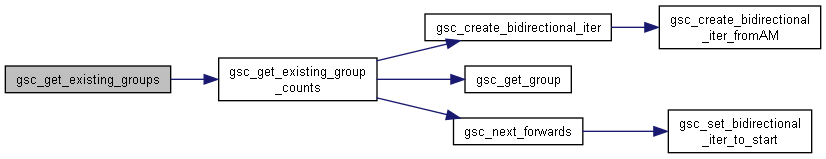
Here is the caller graph for this function:◆ gsc_get_existing_groups()
| size_t gsc_get_existing_groups | ( | gsc_SimData * | d, |
| gsc_GroupNum * | output | ||
| ) |
Identify group numbers that currently have members.
It saves the extant groups to the output vector, and also realigns d->n_groups with the true number of groups.
Frankly this should be deprecated at some point because you can now call gsc_get_existing_group_counts with the output vector for the counts set to NULL and get exactly the same result. (in fact that's the current implementation of this function).
Has short name: get_existing_groups
- Parameters
-
d the gsc_SimData struct on which to perform the operation output Pointer to a location to save the vector of extant groups. It must have at least space for d->n_groups group numbers. Else, if it is NULL, then the group numbers of existing groups are not saved and the only purpose of the function is to return the number of existing groups and update d->n_groups to this number.
- Returns
- The number of entries that have been placed in
output. That is, the number of existing groups.
Definition at line 3539 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_group_bvs()
| unsigned int gsc_get_group_bvs | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| const gsc_EffectID | effID, | ||
| unsigned int | group_size, | ||
| double * | output | ||
| ) |
Gets the breeding values/breeding values/fitnesses of each member of the group.
Has short name: get_group_bvs
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from effID Identifier of the marker effect set to be used to calculate these breeding values group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with the breeding value of each member of the group.
- Returns
- The number of entries of output that have been filled.
Definition at line 4143 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_get_group_genes()
| unsigned int gsc_get_group_genes | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| char ** | output | ||
| ) |
Gets a shallow copy of the genes/alleles of each member of the group.
Not guaranteed to be null-terminated.
Has short name: get_group_genes
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group we want data from group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with pointers to the allele strings of each member of the group. The vector's contents are only shallow copies that should not be freed.
- Returns
- The number of entries of output that have been filled.
Definition at line 3978 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_group_ids()
| unsigned int gsc_get_group_ids | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| gsc_PedigreeID * | output | ||
| ) |
Gets the ids of each member of the group.
Has short name: get_group_ids
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with the pedigree identifiers of each member of the group.
- Returns
- The number of entries of output that have been filled.
Definition at line 4060 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_group_indexes()
| unsigned int gsc_get_group_indexes | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| unsigned int * | output | ||
| ) |
Gets the 0-based global indexes of each member of the group.
Has short name: get_group_indexes
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with the global indexes of each member of the group.
- Returns
- The number of entries of output that have been filled.
Definition at line 4101 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_group_names()
| unsigned int gsc_get_group_names | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| char ** | output | ||
| ) |
Gets a shallow copy of the names of each member of the group.
Has short name: get_group_names
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with pointers to the names of each member of the group. The vector's contents are only shallow copies that should not be freed.
- Returns
- The number of entries of output that have been filled.
Definition at line 4019 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_group_parent_ids()
| unsigned int gsc_get_group_parent_ids | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| const int | whichParent, | ||
| gsc_PedigreeID * | output | ||
| ) |
Gets the ids of either the first or second parent of each member of the group.
Has short name: get_group_parent_ids
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from whichParent 1 to get the first parent of each group member, 2 to get the second. Raises an error and returns -1 (GSC_NA_GLOBALX) if this parameter is not either of those values. group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with the parent's pedigree ID of each member of the group.
- Returns
- The number of entries of output that have been filled.
Definition at line 4181 of file sim-operations.c.
◆ gsc_get_group_parent_names()
| unsigned int gsc_get_group_parent_names | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| const int | whichParent, | ||
| char ** | output | ||
| ) |
Gets the names of either the first or second parent of each member of the group.
Names may be NULL.
Has short name: get_group_parent_names
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from whichParent 1 to get the first parent of each group member, 2 to get the second. Raises an error and returns -1 (GSC_NA_GLOBALX) if this parameter is not either of those values. group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with pointers to the names of each member of the group. The vector's contents are only shallow copies that should not be freed.
- Returns
- The number of entries of output that have been filled.
Definition at line 4232 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_get_group_pedigrees()
| unsigned int gsc_get_group_pedigrees | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| unsigned int | group_size, | ||
| char ** | output | ||
| ) |
Gets the full pedigree string (as per gsc_save_group_full_pedigree() ) of each member of the group.
This function is not fast, or particularly memory-efficient. To avoid recursive string-building/concatenation, this just calls gsc_save_group_full_pedigree() to a temporary file, then reads that file back in to make a list of strings. Contact the package maintainers if you'd benefit from a faster version of this function, otherwise I probably won't bother improving this.
If you just want to have the full pedigrees for further analysis, consider gsc_save_group_full_pedigree(). With the current implementation that one will be faster than this anyway.
Has short name: get_group_pedigrees
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group you want data from group_size the maximum number of values to save to the output vector. Certain values of this parameter (0 and -1/GSC_NA_GLOBALX) give different behaviour for compatibility reasons with past versions of this tool. If group_size has a value of 0 or -1 (GSC_NA_GLOBALX), the function will save data from every member of the group to output. This may be dangerous if the true number of group members is larger than the length of the output vector. output The function will fill this vector with pointers to the names of each member of the group. The vector's contents are heap-allocated and need to be freed.
- Returns
- GSC_NA_GLOBALX if reading/writing to the temporary file failed; otherwise the number of entries of
outputthat have been filled.
Definition at line 4297 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_get_group_size()
| unsigned int gsc_get_group_size | ( | const gsc_SimData * | d, |
| const gsc_GroupNum | group_id | ||
| ) |
Function to count the number of genotypes that currently belong to the specified group.
This goes through and checks every genotype in the gsc_SimData, because there is currently no centralised tracking of groups.
Has short name: get_group_size
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group whose members need to be counted.
- Returns
- the number of genotypes currently belonging to this group. Could be 0.
Definition at line 3940 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function: