For simulation of progression steps in breeding programs. More...
 Collaboration diagram for Crossing and Progression Simulation Functions:
Collaboration diagram for Crossing and Progression Simulation Functions:Modules | |
| Meiosis Simulation Functions | |
| For simulation of meiosis. | |
Functions | |
| gsc_GroupNum | gsc_scaffold_make_new_genotypes (gsc_SimData *d, const gsc_GenOptions g, void *parentIterator, union gsc_datastore_make_genotypes *datastore, int(*parentChooser)(void *, union gsc_datastore_make_genotypes *, unsigned int *, gsc_ParentChoice[static 2]), void(*offspringGenerator)(gsc_SimData *, union gsc_datastore_make_genotypes *, gsc_ParentChoice[static 2], gsc_GenoLocation)) |
| Make new genotypes (generic function) More... | |
| gsc_GroupNum | gsc_make_random_crosses (gsc_SimData *d, const gsc_GroupNum from_group, const unsigned int n_crosses, const unsigned int cap, const gsc_MapID which_map, const gsc_GenOptions g) |
| Performs random crosses among members of a group. More... | |
| gsc_GroupNum | gsc_make_random_crosses_between (gsc_SimData *d, const gsc_GroupNum group1, const gsc_GroupNum group2, const unsigned int n_crosses, const unsigned int cap1, const unsigned int cap2, const gsc_MapID map1, const gsc_MapID map2, const gsc_GenOptions g) |
| Performs random crosses where the first parent comes from one group and the second from another. More... | |
| gsc_GroupNum | gsc_make_targeted_crosses (gsc_SimData *d, const size_t n_combinations, const unsigned int *firstParents, const unsigned int *secondParents, const gsc_MapID map1, const gsc_MapID map2, const gsc_GenOptions g) |
| Performs the crosses of pairs of parents whose indexes are provided in an array. More... | |
| gsc_GroupNum | gsc_self_n_times (gsc_SimData *d, const unsigned int n, const gsc_GroupNum group, const gsc_MapID which_map, const gsc_GenOptions g) |
| Selfs each member of a group for a certain number of generations. More... | |
| gsc_GroupNum | gsc_make_doubled_haploids (gsc_SimData *d, const gsc_GroupNum group, const gsc_MapID which_map, const gsc_GenOptions g) |
| Creates a doubled haploid from each member of a group. More... | |
| gsc_GroupNum | gsc_make_clones (gsc_SimData *d, const gsc_GroupNum group, const _Bool inherit_names, const gsc_GenOptions g) |
| Creates an identical copy of each member of a group. More... | |
| gsc_GroupNum | gsc_make_all_unidirectional_crosses (gsc_SimData *d, const gsc_GroupNum from_group, const gsc_MapID mapID, const gsc_GenOptions g) |
Perform crosses between all pairs of parents in the group from_group and allocates the resulting offspring to a new group. More... | |
| gsc_GroupNum | gsc_make_crosses_from_file (gsc_SimData *d, const char *input_file, const gsc_MapID map1, const gsc_MapID map2, const gsc_GenOptions g) |
| Perform crosses between pairs of parents identified by name in a file and allocate the resulting offspring to a new group. More... | |
| gsc_GroupNum | gsc_make_double_crosses_from_file (gsc_SimData *d, const char *input_file, const gsc_MapID map1, const gsc_MapID map2, const gsc_GenOptions g) |
| Perform crosses between previously-generated offspring of pairs of parents identified by name in a file. More... | |
Detailed Description
For simulation of progression steps in breeding programs.
Function Documentation
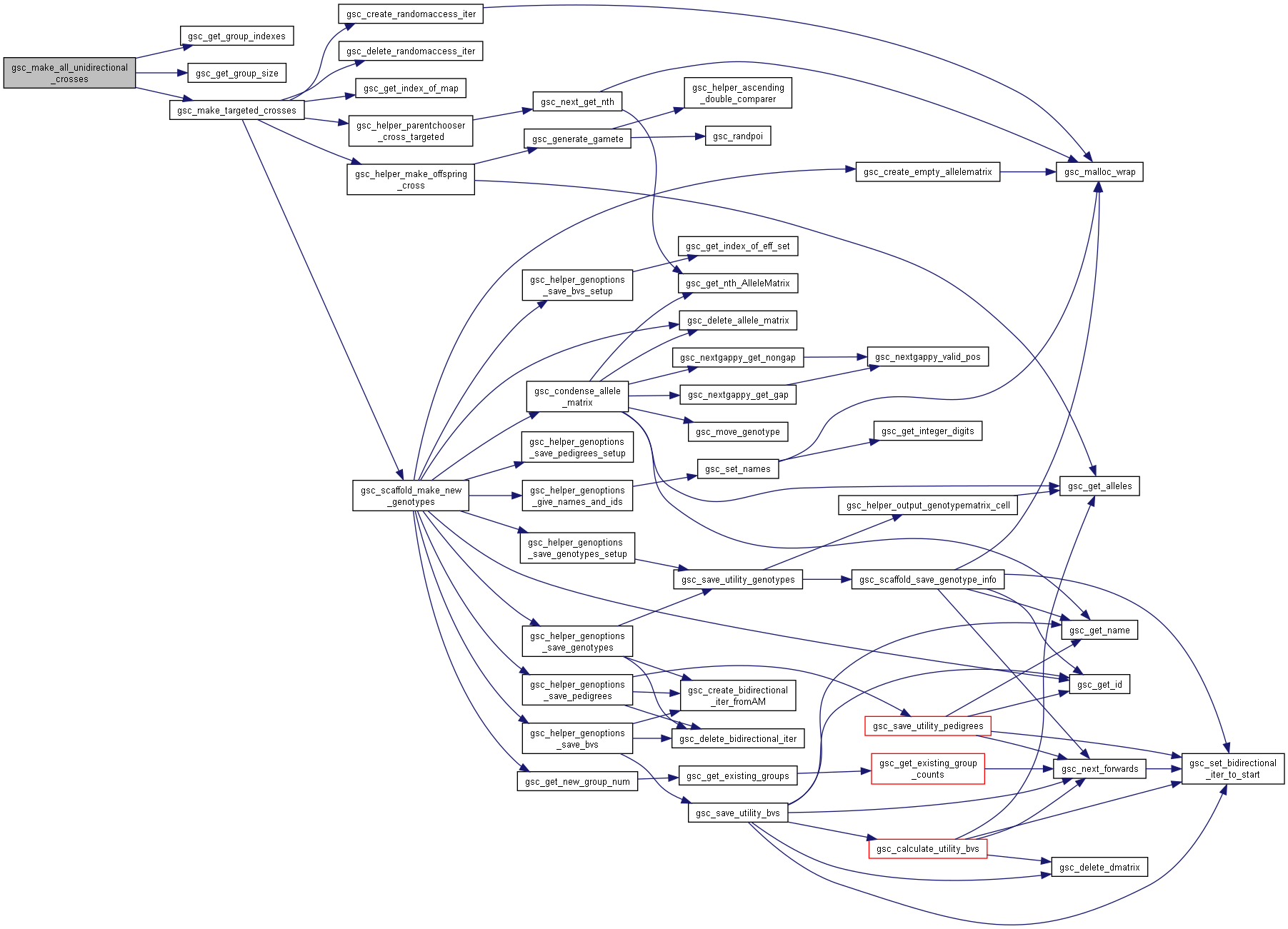
◆ gsc_make_all_unidirectional_crosses()
| gsc_GroupNum gsc_make_all_unidirectional_crosses | ( | gsc_SimData * | d, |
| const gsc_GroupNum | from_group, | ||
| const gsc_MapID | mapID, | ||
| const gsc_GenOptions | g | ||
| ) |
Perform crosses between all pairs of parents in the group from_group and allocates the resulting offspring to a new group.
If the group has n members, there will be $n * (n-1) / 2$ offspring produced.
Preferences in gsc_GenOptions are applied to this cross.
Has short name: make_all_unidirectional_crosses
- Parameters
-
d pointer to the gsc_SimData object containing or markers and parent alleles from_group group number from which to do all these crosses. which_map recombination map to use to generate gametes from members of from_group. If NO_MAP, uses first/primary map by default g options for the gsc_AlleleMatrix created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8783 of file sim-operations.c.
 Here is the call graph for this function:
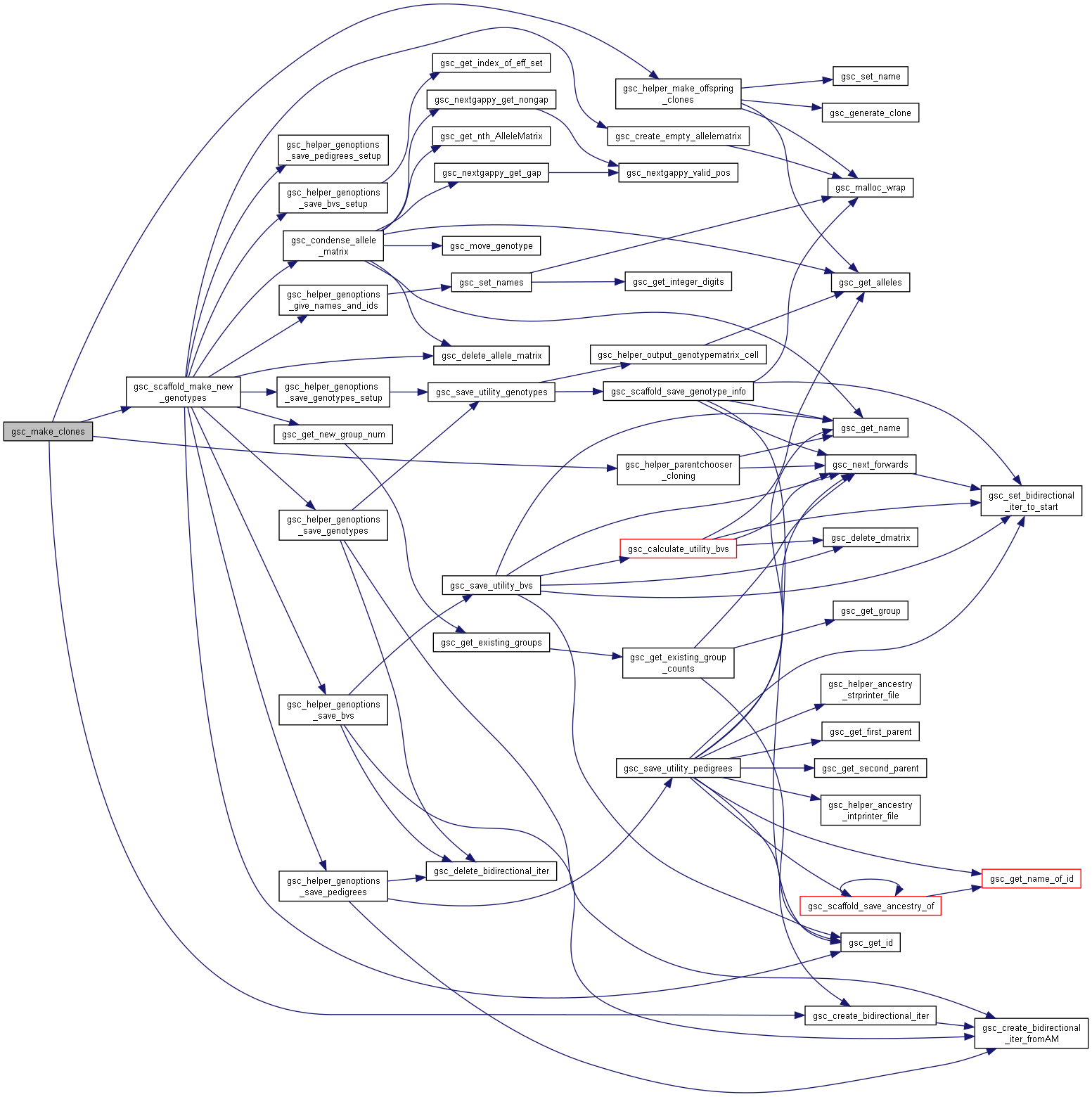
Here is the call graph for this function:◆ gsc_make_clones()
| gsc_GroupNum gsc_make_clones | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const _Bool | inherit_names, | ||
| gsc_GenOptions | g | ||
| ) |
Creates an identical copy of each member of a group.
The resulting genotypes are allocated to a new group.
Preferences in gsc_GenOptions are applied to this operation. The family_size parameter in gsc_GenOptions allows you to generate multiple cloned offspring from each member of the group.
If pedigree tracking and ID allocation are active in gsc_GenOptions, clones are given individual IDs and are children of their single progenitor parent. If the inherit_names parameter is 1/truthy, it overrides whatever naming settings are present in gsc_GenOptions in favour of giving each clone the exact name of the individual it was cloned from.
Clones currently keep the default value for every custom label.
Has short name: make_clones
- Parameters
-
d pointer to the gsc_SimData object that contains the genetic map and genotypes of the parent group. group the genotypes on which to perform the operation. inherit_names true if the clones should have the same name as their parent, false if clone names should be generated according to the GenOptions parameter g options for the genotypes created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8745 of file sim-operations.c.
 Here is the call graph for this function:
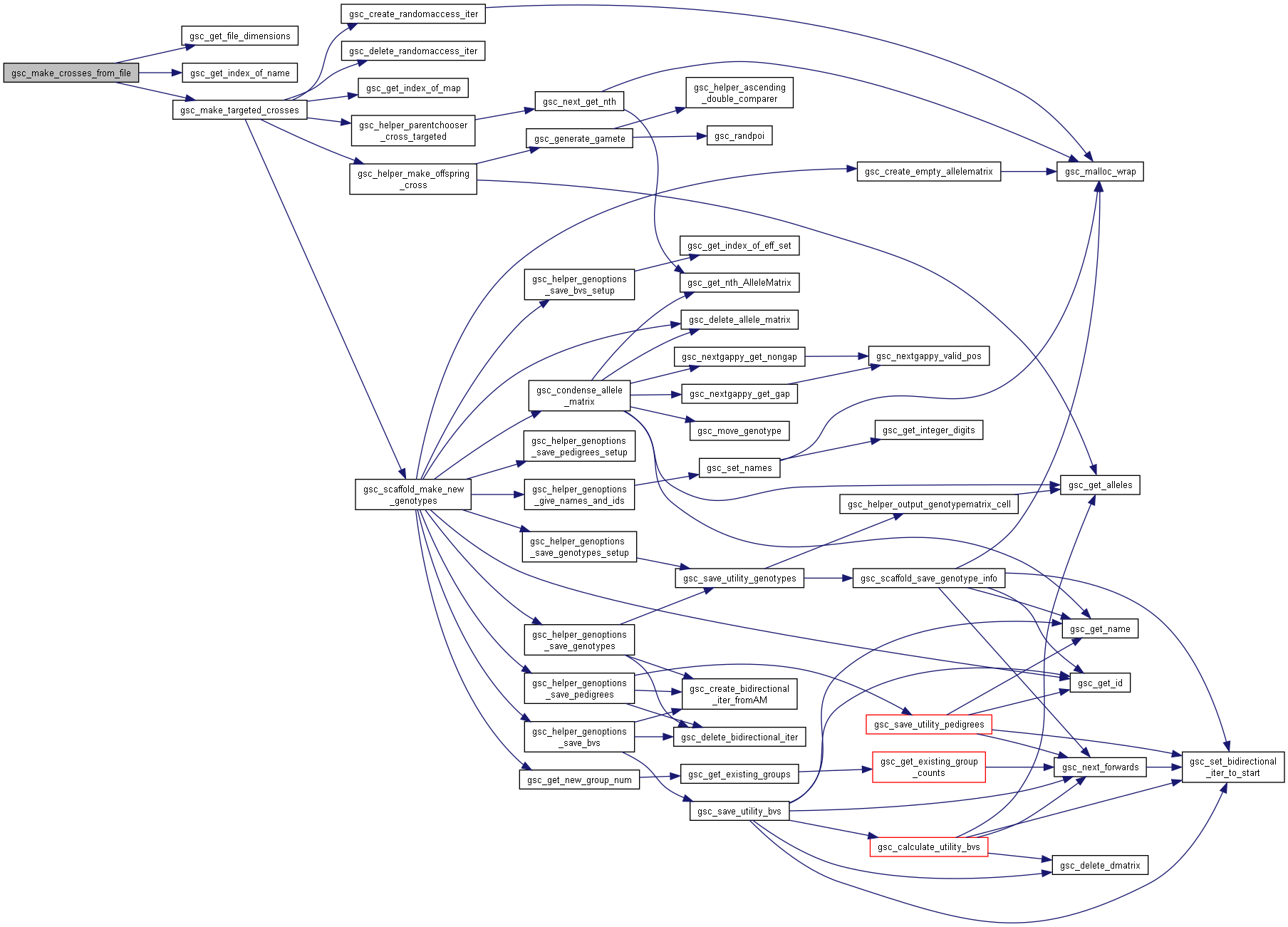
Here is the call graph for this function:◆ gsc_make_crosses_from_file()
| gsc_GroupNum gsc_make_crosses_from_file | ( | gsc_SimData * | d, |
| const char * | input_file, | ||
| const gsc_MapID | map1, | ||
| const gsc_MapID | map2, | ||
| const gsc_GenOptions | g | ||
| ) |
Perform crosses between pairs of parents identified by name in a file and allocate the resulting offspring to a new group.
The input file should have format:
[parent1 name] [parent2 name]
[parent1 name] [parent2 name]
...
where each row represents a separate cross to carry out.
Preferences in gsc_GenOptions are applied to this cross.
Has short name: make_crosses_from_file
- Parameters
-
d pointer to the gsc_SimData object containing or markers and parent alleles input_file file instructing which crosses to perform map1 recombination map to use to generate gametes from the first parent in the crosses in the file. If NO_MAP, uses first/primary map by default map2 recombination map to use to generate gametes from the second parent in the crosses in the file. If NO_MAP, uses first/primary map by default g options for the gsc_AlleleMatrix created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8858 of file sim-operations.c.
 Here is the call graph for this function:
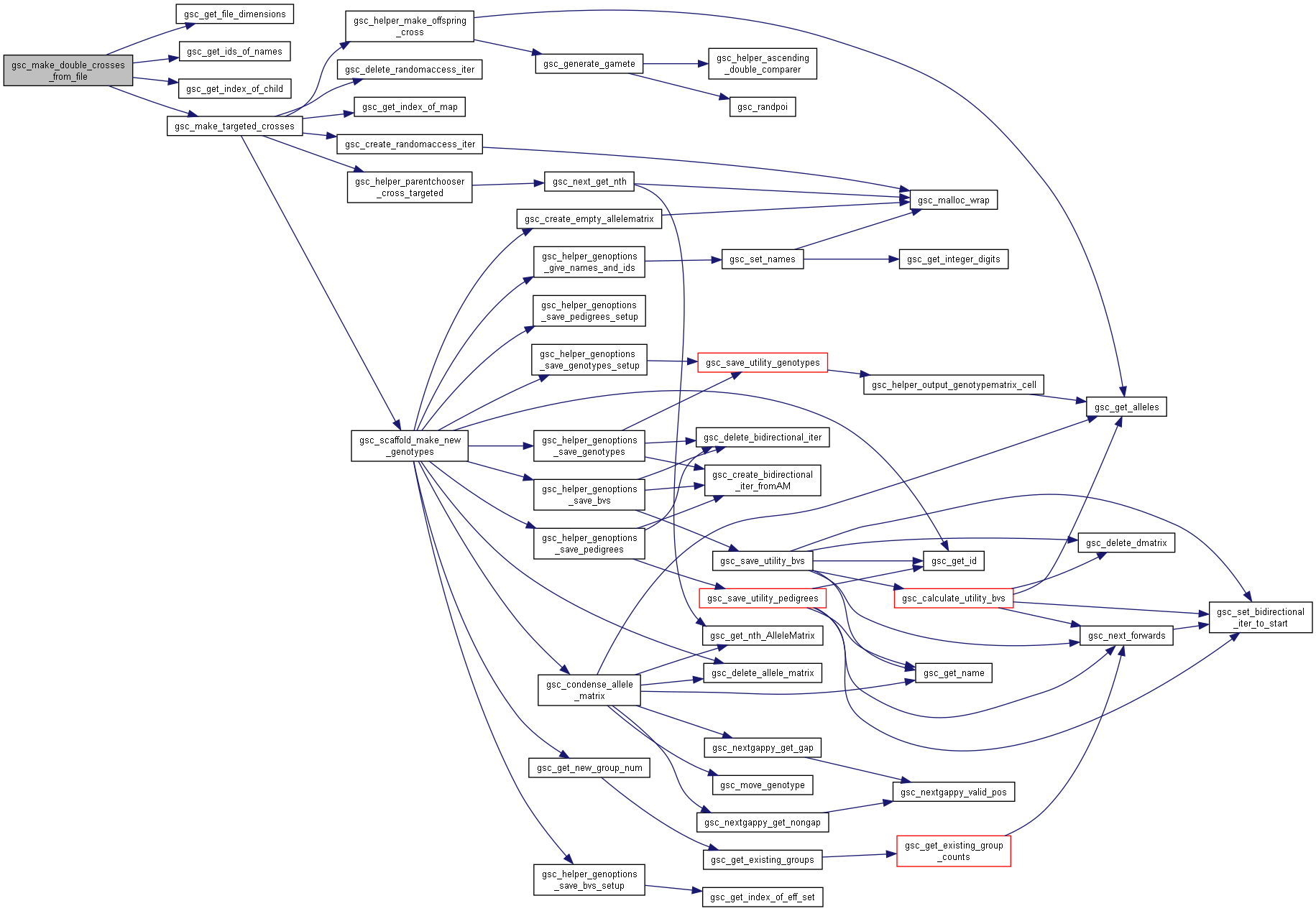
Here is the call graph for this function:◆ gsc_make_double_crosses_from_file()
| gsc_GroupNum gsc_make_double_crosses_from_file | ( | gsc_SimData * | d, |
| const char * | input_file, | ||
| const gsc_MapID | map1, | ||
| const gsc_MapID | map2, | ||
| const gsc_GenOptions | g | ||
| ) |
Perform crosses between previously-generated offspring of pairs of parents identified by name in a file.
The resulting offspring are allocated to a new group.
The input file should have format:
[gparent1 name] [gparent2 name] [gparent3 name] [gparent4 name]
[gparent1 name] [gparent2 name] [gparent3 name] [gparent4 name]
...
where each row represents a separate cross to carry out.
Results of the cross [gparent1 name] x [gparent2 name] and [gparent3 name] x [gparent4 name] must have already been generated with pedigree tracking options turned on. Messages will be printed to stderr if such offspring cannot be found.
Preferences in gsc_GenOptions are applied to this cross.
Has short name: make_double_crosses_from_file
- Parameters
-
d pointer to the gsc_SimData object containing or markers and parent alleles input_file file instructing which crosses to perform map1 recombination map to use to generate gametes from the first parent in each of the crosses in the file. If NO_MAP, uses first/primary map by default map2 recombination map to use to generate gametes from the second parent in each of the crosses in the file. If NO_MAP, uses first/primary map by default g options for the gsc_AlleleMatrix created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8932 of file sim-operations.c.
 Here is the call graph for this function:
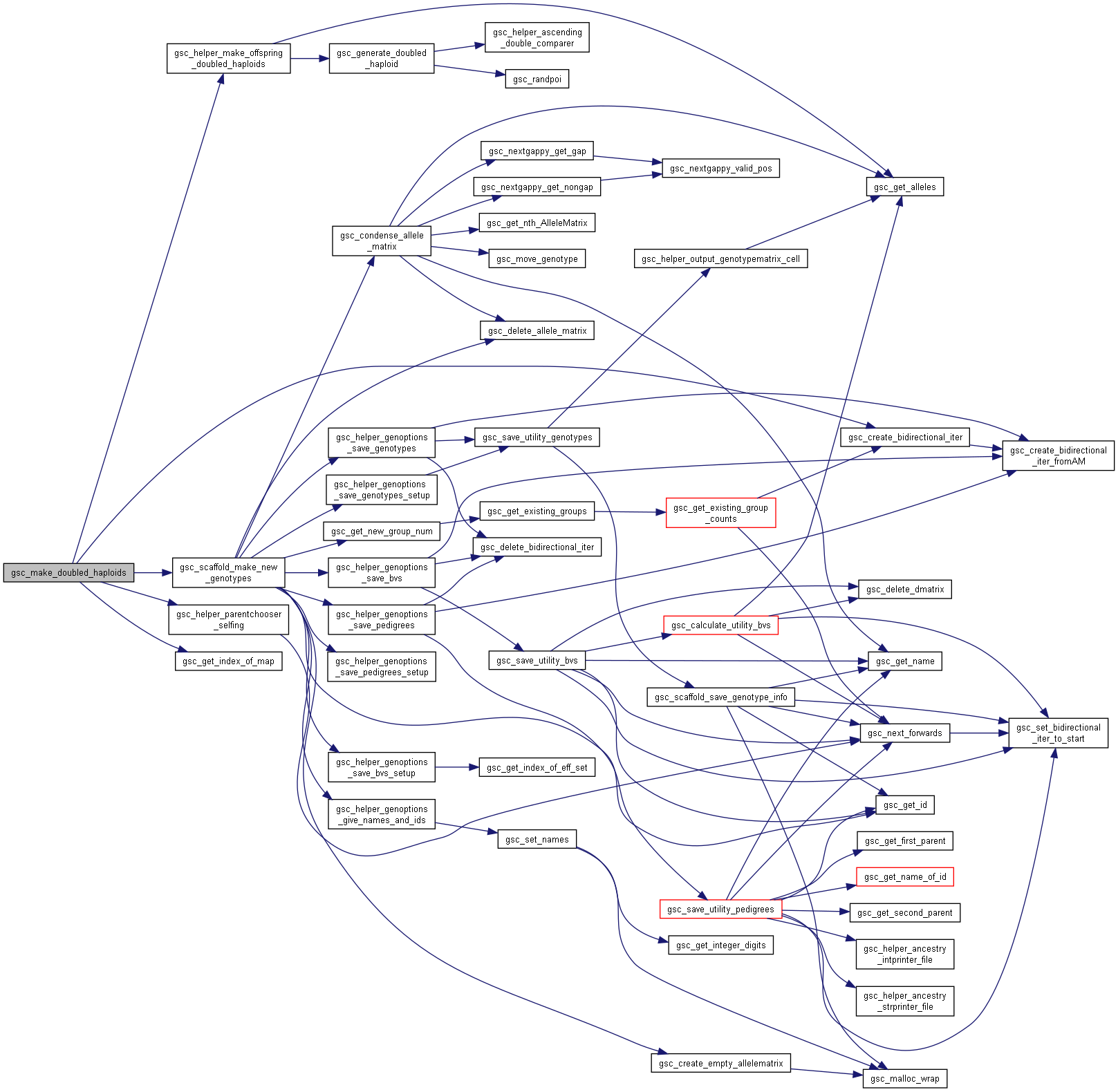
Here is the call graph for this function:◆ gsc_make_doubled_haploids()
| gsc_GroupNum gsc_make_doubled_haploids | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_MapID | which_map, | ||
| const gsc_GenOptions | g | ||
| ) |
Creates a doubled haploid from each member of a group.
The resulting genotypes are allocated to a new group.
Preferences in gsc_GenOptions are applied to this operation. The family_size parameter in gsc_GenOptions allows you to generate multiple doubled haploid offspring from each member of the group. These multiple offspring all originate from independent processes of generating a gamete then doubling its alleles.
Has short name: make_doubled_haploids
- Parameters
-
d pointer to the gsc_SimData object that contains the genetic map and genotypes of the parent group. group the genotypes on which to perform the operation. which_map recombination map to use to generate gametes from members of group. If NO_MAP, uses first/primary map by default g options for the genotypes created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8637 of file sim-operations.c.
 Here is the call graph for this function:
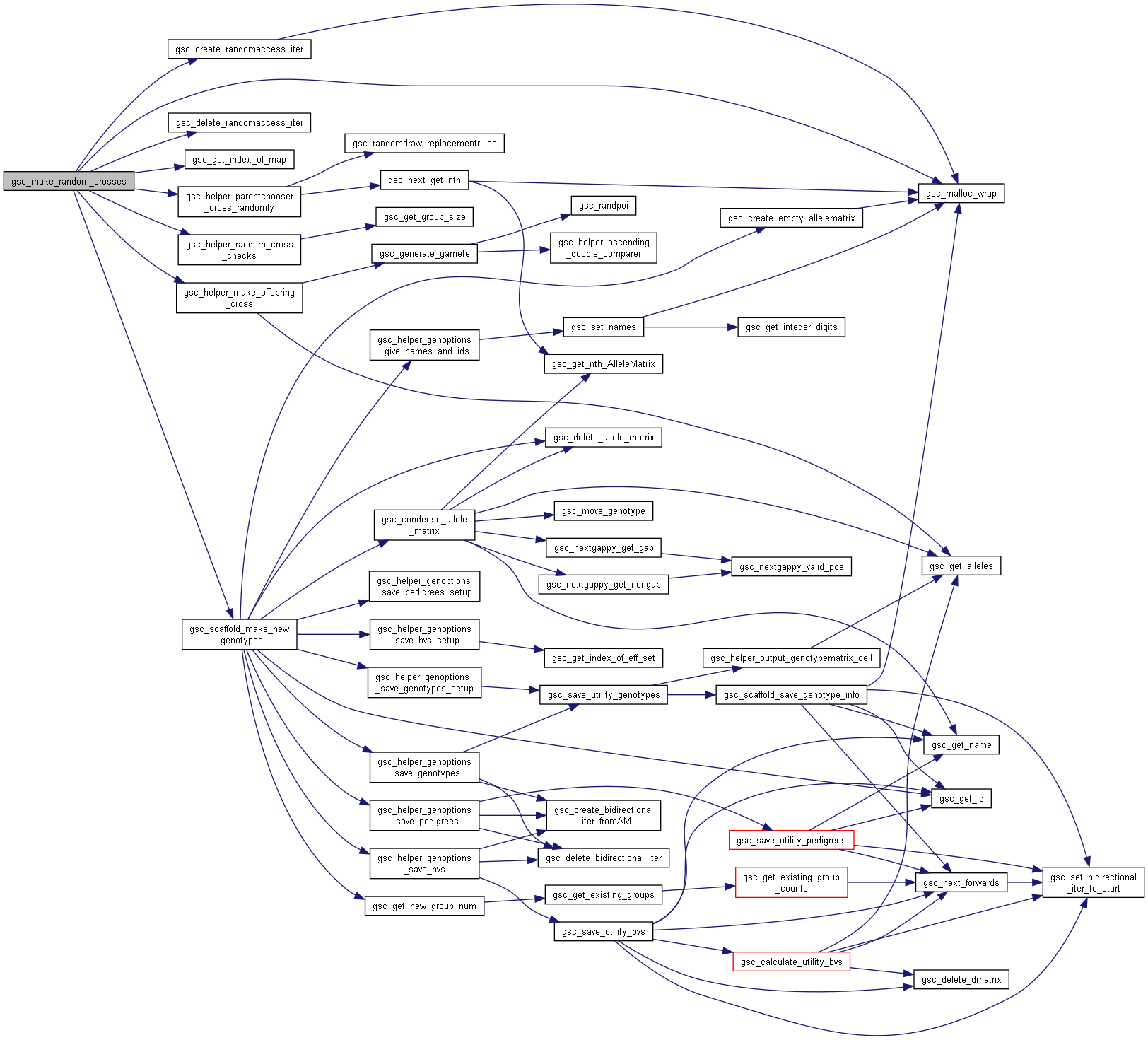
Here is the call graph for this function:◆ gsc_make_random_crosses()
| gsc_GroupNum gsc_make_random_crosses | ( | gsc_SimData * | d, |
| const gsc_GroupNum | from_group, | ||
| const unsigned int | n_crosses, | ||
| const unsigned int | cap, | ||
| const gsc_MapID | which_map, | ||
| const gsc_GenOptions | g | ||
| ) |
Performs random crosses among members of a group.
The group must have at least two members. Selfing is not considered a form of random crossing. The resulting genotypes are allocated to a new group.
Preferences in gsc_GenOptions are applied to this cross. The family_size parameter in gsc_GenOptions allows you to repeat each particular randomly-chosen cross a certain number of times.
Parents are drawn uniformly from the group when picking which crosses to make.
Has short name: make_random_crosses
- Parameters
-
d pointer to the gsc_SimData object that contains the genetic map and genotypes of the parent group. from_group group number from which to draw the parents. cap If set, the maximum number of times each member of from_group can be used as the parent of a cross. Set to 0 for no restriction on the number of offspring produced by a given member of from_group n_crosses number of random pairs of parents to cross. which_map recombination map to use to generate gametes from members of from_group. If NO_MAP, uses first/primary map by default g options for the genotypes created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8093 of file sim-operations.c.
 Here is the call graph for this function:
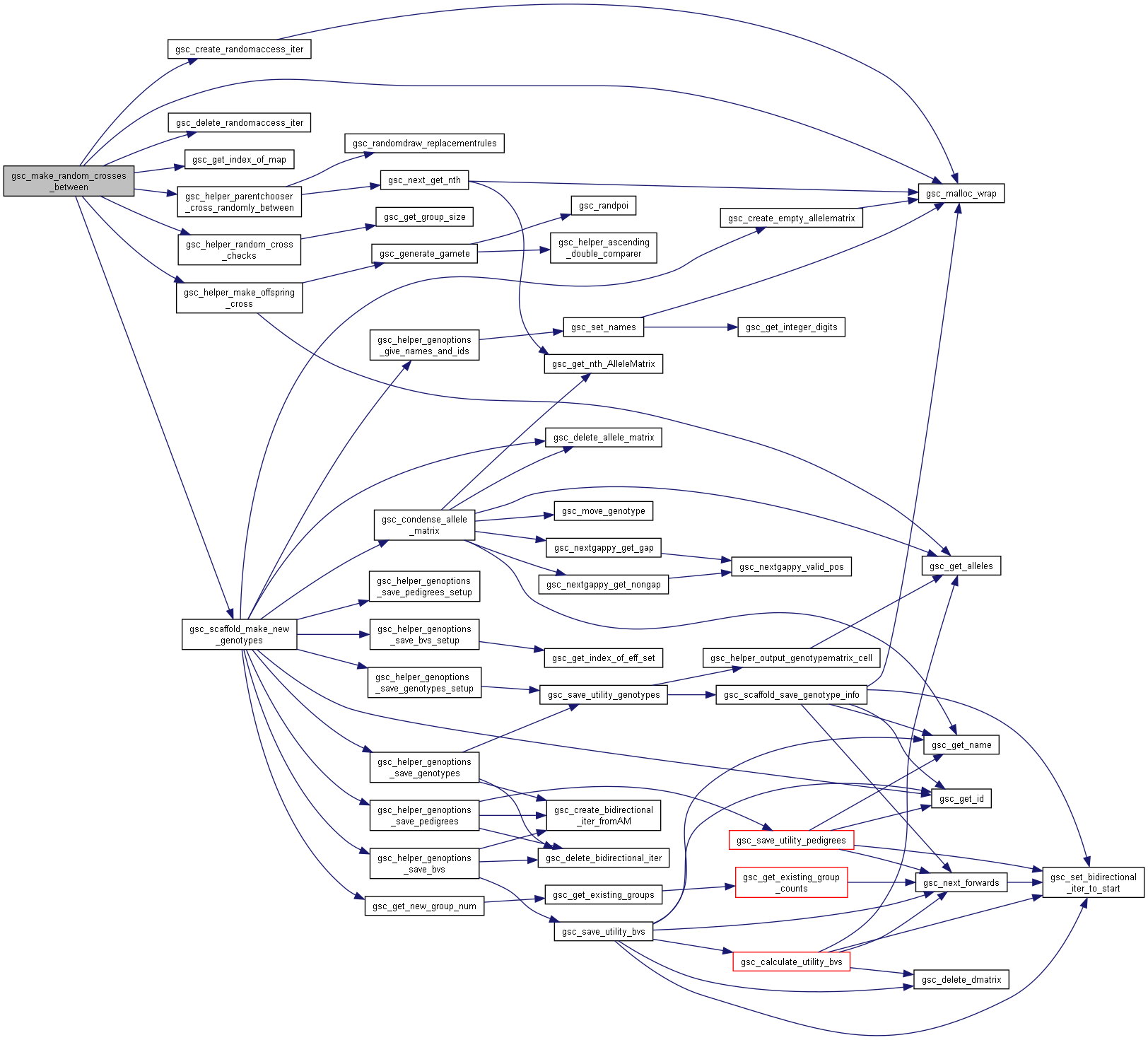
Here is the call graph for this function:◆ gsc_make_random_crosses_between()
| gsc_GroupNum gsc_make_random_crosses_between | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group1, | ||
| const gsc_GroupNum | group2, | ||
| const unsigned int | n_crosses, | ||
| const unsigned int | cap1, | ||
| const unsigned int | cap2, | ||
| const gsc_MapID | map1, | ||
| const gsc_MapID | map2, | ||
| const gsc_GenOptions | g | ||
| ) |
Performs random crosses where the first parent comes from one group and the second from another.
The group must have at least two members. The resulting genotypes are allocated to a new group.
Preferences in gsc_GenOptions are applied to this cross. The family_size parameter in gsc_GenOptions allows you to repeat each particular randomly-chosen cross a certain number of times.
Parents are drawn uniformly from the group when picking which crosses to make.
Parameters set_parent_gp1 and set_parent_gp2 are deprecated and removed! Use gsc_make_group_from and gsc_combine_groups to temporarily move an individual to their own group if you wish to cross randomly from a group to an individual.
Has short name: make_random_crosses_between
- Parameters
-
d pointer to the gsc_SimData object that contains the genetic map and genotypes of the parent group. group1 group number from which to draw the first parent. group2 group number from which to draw the second parent. n_crosses number of random pairs of parents to cross. cap1 If set, the maximum number of times each member of group1 can be used as the parent of a cross. Set to 0 for no restriction on the number of offspring produced by a given member of group1 cap2 If set, the maximum number of times each member of group2 can be used as the parent of a cross. Set to 0 for no restriction on the number of offspring produced by a given member of group2 map1 recombination map to use to generate gametes from members of group1. If NO_MAP, uses first/primary map by default map2 recombination map to use to generate gametes from members of group2. If NO_MAP, uses first/primary map by default g options for the genotypes created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8279 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_make_targeted_crosses()
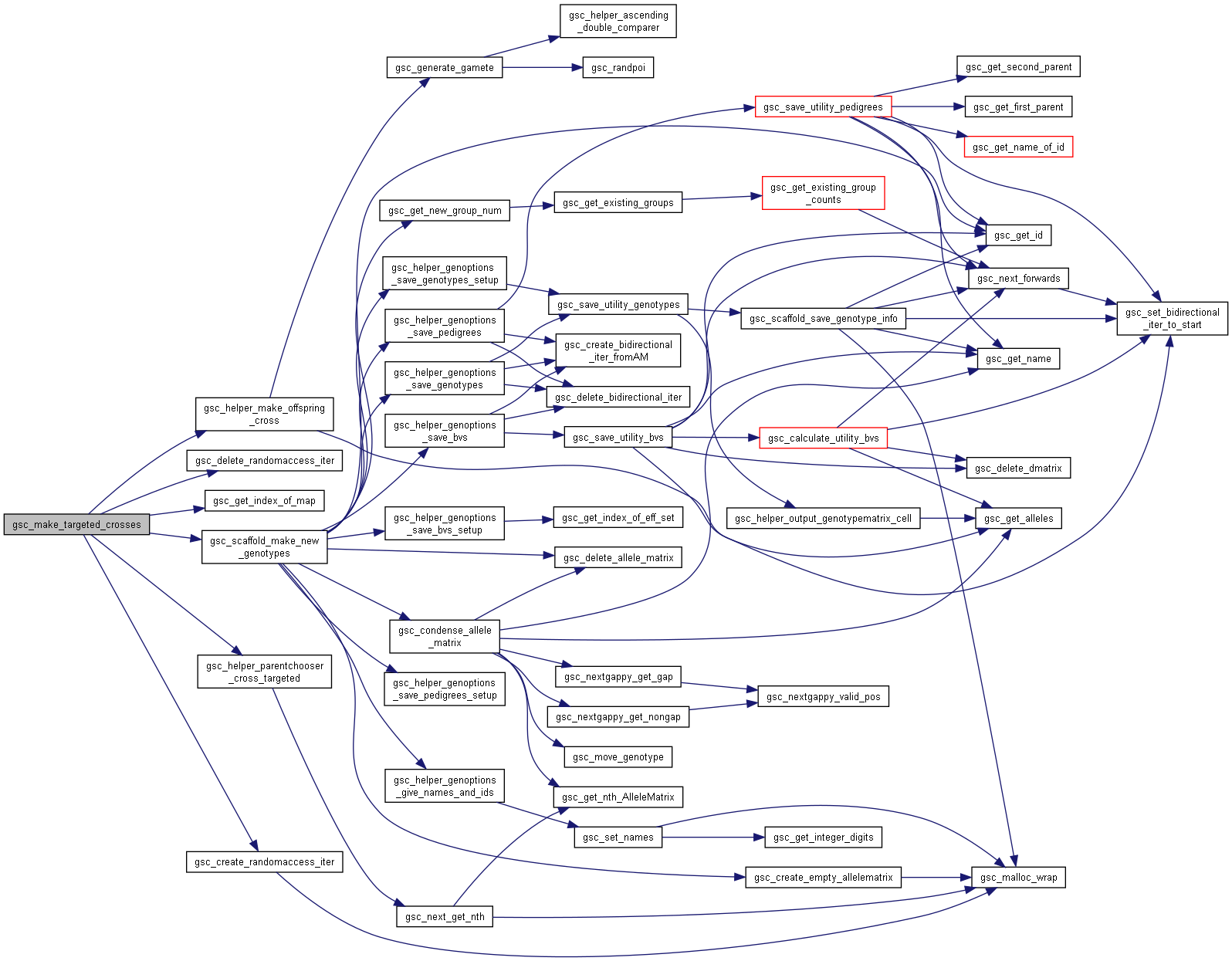
| gsc_GroupNum gsc_make_targeted_crosses | ( | gsc_SimData * | d, |
| const size_t | n_combinations, | ||
| const unsigned int * | firstParents, | ||
| const unsigned int * | secondParents, | ||
| const gsc_MapID | map1, | ||
| const gsc_MapID | map2, | ||
| const gsc_GenOptions | g | ||
| ) |
Performs the crosses of pairs of parents whose indexes are provided in an array.
The resulting genotypes are allocated to a new group.
Preferences in gsc_GenOptions are applied to this cross. The family_size parameter in gsc_GenOptions allows you to repeat each particular cross a certain number of times.
Previously had a parameter combinations[2][n_combinations] instead of firstParents and secondParents. This was changed to lower the boilerplate needs of calling this function: now there is no need to create a 2-wide int* to hold the two separate parent vectors if they already exist.
Has short name: make_targeted_crosses
- Parameters
-
d pointer to the gsc_SimData object that includes genetic map data and allele data needed to simulate crossing. n_combinations the number of pairs to cross. firstParents a vector of indexes of parents to be the first parent of each cross. The vector must have at least [n_combinations] entries. secondParents a vector of indexes of parents to be the second parent of each cross. The vector must have at least [n_combinations] entries. firstParents[0] is crossed to secondParents[0], firstParents[1] is crossed to secondParents[1], and so forth. map1 recombination map to use to generate gametes from firstParents. If NO_MAP, uses first/primary map by default map2 recombination map to use to generate gametes from secondParents. If NO_MAP, uses first/primary map by default g options for the genotypes created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8420 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_scaffold_make_new_genotypes()
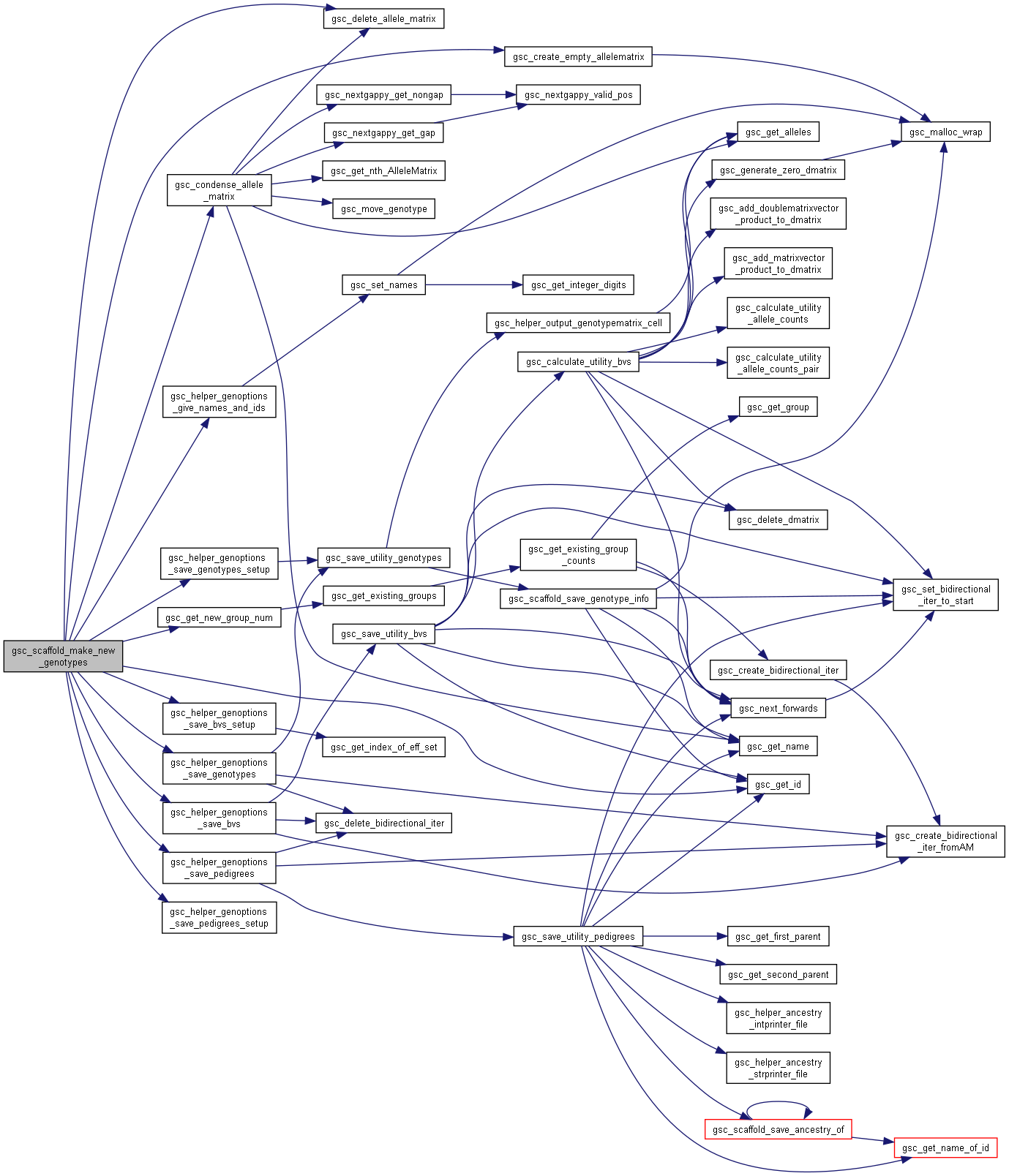
| gsc_GroupNum gsc_scaffold_make_new_genotypes | ( | gsc_SimData * | d, |
| const gsc_GenOptions | g, | ||
| void * | parentIterator, | ||
| union gsc_datastore_make_genotypes * | datastore, | ||
| int(*)(void *, union gsc_datastore_make_genotypes *, unsigned int *, gsc_ParentChoice[static 2]) | parentChooser, | ||
| void(*)(gsc_SimData *, union gsc_datastore_make_genotypes *, gsc_ParentChoice[static 2], gsc_GenoLocation) | offspringGenerator | ||
| ) |
Make new genotypes (generic function)
Applies all settings from gsc_GenOptions g.
Takes two parameter functions: parentChooser and offspringGenerator, described below.
parentChooser should function as a 'generator' of parents for the new genotypes. Every time the function is called, it should return a truthy value and save the gsc_GenoLocations of its chosen parents in its final parameter, or return a falsy value if there are no more offspring to be created. It has access to three informational parameters to help it choose parents: in order: parentIterator, passed by the calling function, datastore, passed by the calling function, and a pointer to a counter representing the number of times parentChooser has been called by this function prior to the current call. This generic function trusts that parentChooser will not return a truthy value if setting the parent choice gsc_GenoLocations to invalid values (of a type that cannot be handled by the corresponding offspringGenerator).
offspringGenerator should make the call that generates alleles for a new genotype and saves them to a given gsc_GenoLocation. (It may also make any other necessary specific modifications to other data about the new genotype). It will be called g.family_size times for each truthy return value of parentChooser. As parameters, it receives a pointer to the gsc_SimData, and datastore as passed by the calling function and potentially modified by parentChooser, the gsc_GenoLocations of up to two parents, as set by parentChooser, and the location to which it is to save the new genotype.
- Returns
- group number of new group created, or GSC_NO_GROUP if no group was created or the new group was empty.
Definition at line 7868 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_self_n_times()
| gsc_GroupNum gsc_self_n_times | ( | gsc_SimData * | d, |
| const unsigned int | n, | ||
| const gsc_GroupNum | group, | ||
| const gsc_MapID | which_map, | ||
| const gsc_GenOptions | g | ||
| ) |
Selfs each member of a group for a certain number of generations.
The resulting genotypes are allocated to a new group.
Only the genotype after all n generations is saved. Intermediate steps will be lost.
Preferences in gsc_GenOptions are applied to this operation. The family_size parameter in gsc_GenOptions allows you to generate multiple selfed offspring from each member of the group. These multiple selfed offspring all originate from independent processes of selfing the parent with itself then selfing that intermediate offspring with itself for the remaining n-1 generations.
Has short name: self_n_times
- Parameters
-
d pointer to the gsc_SimData object that contains the genetic map and genotypes of the parent group. n number of generations of selfing simulation to carry out. group the genotypes on which to perform these n generations of selfing. which_map recombination map to use to generate gametes from members of group. If NO_MAP, uses first/primary map by default g options for the genotypes created.
- See also
- gsc_GenOptions
- Returns
- the group number of the group to which the produced offspring were allocated.
Definition at line 8561 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: