For simulation of selection or structure in breeding programs. More...
Functions | |
| gsc_GroupNum | gsc_combine_groups (gsc_SimData *d, const size_t list_len, const gsc_GroupNum *grouplist) |
| Combine a set of groups into one group. More... | |
| gsc_GroupNum | gsc_make_group_from (gsc_SimData *d, const size_t index_list_len, const unsigned int *genotype_indexes) |
| Take a list of indexes and allocate the genotypes at those indexes to a new group. More... | |
| gsc_GroupNum | gsc_split_by_label_value (gsc_SimData *d, const gsc_GroupNum group, const gsc_LabelID whichLabel, const int valueToSplit) |
| Allocates the genotypes with a particular value of a label to a new group. More... | |
| gsc_GroupNum | gsc_split_by_label_range (gsc_SimData *d, const gsc_GroupNum group, const gsc_LabelID whichLabel, const int valueLowBound, const int valueHighBound) |
| Allocates the genotypes with values of a label in a particular range to a new group. More... | |
| size_t | gsc_scaffold_split_by_somequality (gsc_SimData *d, const gsc_GroupNum group_id, void *somequality_data, gsc_GroupNum(*somequality_tester)(gsc_GenoLocation, void *, size_t, size_t, gsc_GroupNum *), size_t maxentries_results, gsc_GroupNum *results) |
| Split by some quality (generic function) More... | |
| size_t | gsc_split_into_individuals (gsc_SimData *d, const gsc_GroupNum group_id, size_t maxentries_results, gsc_GroupNum *results) |
| Split a group into n one-member groups. More... | |
| size_t | gsc_split_into_families (gsc_SimData *d, const gsc_GroupNum group_id, size_t maxentries_results, gsc_GroupNum *results) |
| Split a group into families by their pedigrees. More... | |
| size_t | gsc_split_into_halfsib_families (gsc_SimData *d, const gsc_GroupNum group_id, const int parent, size_t maxentries_results, gsc_GroupNum *results) |
| Split a group into families of half-siblings by shared first or second parent. More... | |
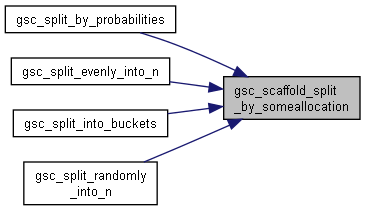
| size_t | gsc_scaffold_split_by_someallocation (gsc_SimData *d, const gsc_GroupNum group_id, void *someallocator_data, gsc_GroupNum(*someallocator)(gsc_GenoLocation, gsc_SimData *, void *, size_t, size_t *, gsc_GroupNum *), size_t n_outgroups, gsc_GroupNum *outgroups) |
| Split by some allocator (generic function) More... | |
| gsc_GroupNum | gsc_split_evenly_into_two (gsc_SimData *d, const gsc_GroupNum group_id) |
| Split a group into two groups of equal size (or size differing only by one, if the original group had an odd number of members) using a random permutation of the group members to determine which goes where. More... | |
| size_t | gsc_split_evenly_into_n (gsc_SimData *d, const gsc_GroupNum group_id, const size_t n, gsc_GroupNum *results) |
| Split a group into n groups of equal size (or size differing only by one, if n does not perfectly divide the group size.), using a random permutation of the group members to determine which goes where. More... | |
| size_t | gsc_split_into_buckets (gsc_SimData *d, const gsc_GroupNum group_id, const size_t n, const unsigned int *counts, gsc_GroupNum *results) |
| Split a group into n groups of equal size (or size differing only by one, if n does not perfectly divide the group size), using a random permutation of the group members to determine which goes where. More... | |
| gsc_GroupNum | gsc_split_randomly_into_two (gsc_SimData *d, const gsc_GroupNum group_id) |
| Flip a coin for each member of the group to decide if it should be moved to the new group. More... | |
| size_t | gsc_split_randomly_into_n (gsc_SimData *d, const gsc_GroupNum group_id, const size_t n, gsc_GroupNum *results) |
| Allocate each member of the group to one of n groups with equal probability. More... | |
| size_t | gsc_split_by_probabilities (gsc_SimData *d, const gsc_GroupNum group_id, const size_t n, const double *probs, gsc_GroupNum *results) |
| Allocate each member of the group to one of n groups with custom probabilities for each group. More... | |
Detailed Description
For simulation of selection or structure in breeding programs.
Function Documentation
◆ gsc_combine_groups()
| gsc_GroupNum gsc_combine_groups | ( | gsc_SimData * | d, |
| const size_t | list_len, | ||
| const gsc_GroupNum * | grouplist | ||
| ) |
Combine a set of groups into one group.
The function does so by setting the group membership of every genotype belonging to one of the groups to the same group number.
Has short name: combine_groups
- Parameters
-
d the gsc_SimData struct on which to perform the operation list_len the number of groups to be combined grouplist an array of at least [list_len] group numbers, representing the groups that are to be combined.
- Returns
- the group number of the new combined group.
Definition at line 2459 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_make_group_from()
| gsc_GroupNum gsc_make_group_from | ( | gsc_SimData * | d, |
| const size_t | index_list_len, | ||
| const unsigned int * | genotype_indexes | ||
| ) |
Take a list of indexes and allocate the genotypes at those indexes to a new group.
Does not check if all the indexes are valid/if all indexes have successfully had their groups changed.
Has short name: make_group_from
- Parameters
-
d the gsc_SimData struct on which to perform the operation index_list_len the number of indexes provided genotype_indexes an array containing the global indexes (0-based, starting at the first entry at d->m) of the genotypes to allocate to the new group. This array must have at least [index_list_len] entries.
- Returns
- the group number of the new group to which the provided indexes have been allocated.
Definition at line 2565 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_scaffold_split_by_someallocation()
| size_t gsc_scaffold_split_by_someallocation | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| void * | someallocator_data, | ||
| gsc_GroupNum(*)(gsc_GenoLocation, gsc_SimData *, void *, size_t, size_t *, gsc_GroupNum *) | someallocator, | ||
| size_t | n_outgroups, | ||
| gsc_GroupNum * | outgroups | ||
| ) |
Split by some allocator (generic function)
Allocator: you know how many groups you're splitting into from the beginning.
Takes a parameter someallocator that uses a gsc_GenoLocation, gsc_SimData, someallocator_data, the total number of groups you're splitting into, the current number of new groups that have allocations (as a pointer so someallocator can modify it to hold the number of subgroups that have been found, which will become the return value of this function), and the list of group numbers of new groups. It should return a group number (taken from some position in that final parameter) if the genotype at that gsc_GenoLocation is to be added to one of the groups that already has allocations, or return GSC_NO_GROUP if allocation fails. If allocation fails the genotype will remain in the original group.
- Returns
- number of groups created. May be n_outgroups or less.
Definition at line 3106 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_scaffold_split_by_somequality()
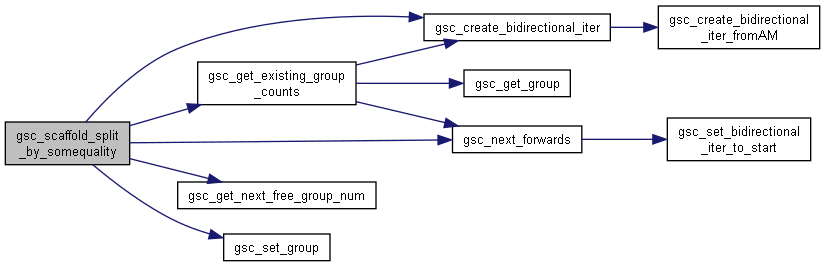
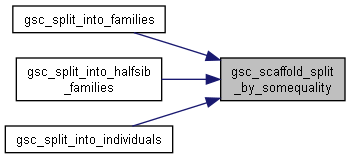
| size_t gsc_scaffold_split_by_somequality | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| void * | somequality_data, | ||
| gsc_GroupNum(*)(gsc_GenoLocation, void *, size_t, size_t, gsc_GroupNum *) | somequality_tester, | ||
| size_t | maxentries_results, | ||
| gsc_GroupNum * | results | ||
| ) |
Split by some quality (generic function)
Somequality: you don't know how many variants of that quality there are, so you don't initially know how many groups you will have.
Takes a parameter somequality_tester that uses a gsc_GenoLocation, somequality_data, the maximum possible number of groups you're splitting into, the current number of new groups that have allocations, and the list of potential group numbers for new groups. It should return a group number (taken from some position in that final parameter) if the genotype at that gsc_GenoLocation is to be added to one of the groups that already has allocations, or return GSC_NO_GROUP if it is to be allocated to one of the groups beyond the [value of the second-last parameter]th (This is so that this caller function can keep allocate new group numbers and keep track of how many subgroups have been found).
If the number of variants ends up being larger than maxentries_results, further variants will still be allocated to new groups, but will not saved to results. The calling function can know this is the case if the returned value is larger than the parameter maxentries_results.
- Returns
- number of groups created. May be less or more than maxentries_results.
Definition at line 2733 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
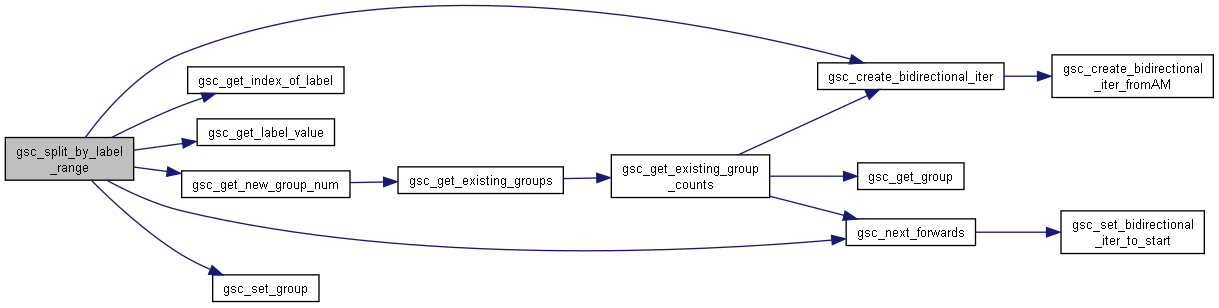
Here is the caller graph for this function:◆ gsc_split_by_label_range()
| gsc_GroupNum gsc_split_by_label_range | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_LabelID | whichLabel, | ||
| const int | valueLowBound, | ||
| const int | valueHighBound | ||
| ) |
Allocates the genotypes with values of a label in a particular range to a new group.
Searches through every genotype (or every member of the group, if a specific group is given) to find all genotypes with value in the whichLabelth label between valueLowBound and valueHighBound inclusive, and puts those genotypes into a new group.
Returns 0 for invalid parameters, or if no genotypes were found that fit the criteria to be moved to the new group.
Has short name: split_by_label_range
- Parameters
-
d the gsc_SimData struct on which to perform the operation group If group> 0, then only genotypes with group numbergroupANDwhichLabelvaluevalueToSplitwill be moved to the new group. Otherwise, all genotypes withwhichLabelvaluevalueToSplitwill be moved to the next group.whichLabel the label id of the relevant label. valueLowBound the minimum value of the label for the genotypes that will be moved to the new group. valueHighBound the maximum value of the label for the genotypes that will be moved to the new group.
- Returns
- the group number of the new group to which the genotypes with that value for that label were allocated, or 0 if no genotypes that fit the criteria were found.
Definition at line 2673 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_by_label_value()
| gsc_GroupNum gsc_split_by_label_value | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group, | ||
| const gsc_LabelID | whichLabel, | ||
| const int | valueToSplit | ||
| ) |
Allocates the genotypes with a particular value of a label to a new group.
Searches through every genotype (or every member of the group, if a specific group is given) to find all genotypes with value valueToSplit in the whichLabelth label, and puts those genotypes into a new group.
Returns 0 for invalid parameters, or if no genotypes were found that fit the criteria to be moved to the new group.
Has short name: split_by_label_value
- Parameters
-
d the gsc_SimData struct on which to perform the operation group If group> 0, then only genotypes with group numbergroupANDwhichLabelvaluevalueToSplitwill be moved to the new group. Otherwise, all genotypes withwhichLabelvaluevalueToSplitwill be moved to the next group.whichLabel the label id of the relevant label. valueToSplit the value of the label that defines the genotypes that will be moved to the new group.
- Returns
- the group number of the new group to which the genotypes with that value for that label were allocated, or 0 if no genotypes that fit the criteria were found.
Definition at line 2617 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_by_probabilities()
| size_t gsc_split_by_probabilities | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| const size_t | n, | ||
| const double * | probs, | ||
| gsc_GroupNum * | results | ||
| ) |
Allocate each member of the group to one of n groups with custom probabilities for each group.
There is no guarantee that all groups will have members. There is no guarantee the groups will be near the same size.
The probability of staying in the old group (group_id) is probs[0]. The probability of going to the first new group is probs[1], etc.. The probability of going to the nth group is 1 minus the sum of all probabilities in probs.
The function draws from a uniform distribution, then uses cumulative sums to determine to which group the group member is allocated. If the sum of the probabilities in probs adds up to more than 1, a warning is raised, and the group numbers for which the cumulative sum of probs is greater than 1 have no chance of being allocated members.
Has short name: split_by_probabilities
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split n the number of groups among which to randomly distribute group members. probs pointer to an array of length n-1 containing the probability of being allocated to each group. The probability of going to the last group is 1 - sum(probs). results NULL if the caller does not care to know the identifiers of the groups created, or a pointer to an array to which these identifiers should be saved. It is assumed that the array is long enough to store n identifiers.
- Returns
- the number of groups created by this process.
Definition at line 3473 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_evenly_into_n()
| size_t gsc_split_evenly_into_n | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| const size_t | n, | ||
| gsc_GroupNum * | results | ||
| ) |
Split a group into n groups of equal size (or size differing only by one, if n does not perfectly divide the group size.), using a random permutation of the group members to determine which goes where.
Of the split groups produced, the first has the same group number as the original group (parameter group_id).
A more general approach to this task: gsc_split_into_buckets()
Has short name: split_evenly_into_n
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split n the number of groups among which to randomly distribute group members. results NULL if the caller does not care to know the identifiers of the groups created, or a pointer to an array to which these identifiers should be saved. It is assumed that the array is long enough to store n identifiers.
- Returns
- the number of groups created by this process.
Definition at line 3197 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_evenly_into_two()
| gsc_GroupNum gsc_split_evenly_into_two | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id | ||
| ) |
Split a group into two groups of equal size (or size differing only by one, if the original group had an odd number of members) using a random permutation of the group members to determine which goes where.
Of the two groups produced, one has the same group number as the original group (parameter group_id) and the other has the return value as its group number.
If the original group size was odd, the new group/the return value will have the slightly smaller size.
A more general approach to this task: gsc_split_evenly_into_n()
An alternate approach to splitting a group in two: gsc_split_randomly_into_two()
Has short name: split_evenly_into_two
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split
- Returns
- the group number of the new group to which half the members of the old group have been allocated.
Definition at line 3051 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_into_buckets()
| size_t gsc_split_into_buckets | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| const size_t | n, | ||
| const unsigned int * | counts, | ||
| gsc_GroupNum * | results | ||
| ) |
Split a group into n groups of equal size (or size differing only by one, if n does not perfectly divide the group size), using a random permutation of the group members to determine which goes where.
Of the split groups produced, the first has the same group number as the original group (parameter group_id).
The number of members staying in the old group (group_id) is counts[0]. The number going to the first new group is counts[1], etc.. The number going to the nth group is group_id's group size - sum(counts).
The function calculates a random permutation of the group members, then uses cumulative sums to determine to which group the group member is allocated. If the sum of the desired group sizes adds up to more than the number of group members, a warning is raised, and the group numbers for which the cumulative sum of counts is greater than the group size will not be allocated members. That is, the group capacities are filled from first to last, leaving later groups unfilled if there are not enough group members to occupy all capacities.
Has short name: split_into_buckets
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split n the number of groups among which to randomly distribute group members. counts pointer to an array of length at least n-1 containing the number of members to allocate to each group. The number of members in the last group is group_id's group size - sum(counts). results NULL if the caller does not care to know the identifiers of the groups created, or a pointer to an array to which these identifiers should be saved. It is assumed that the array is long enough to store n identifiers.
- Returns
- the number of groups created by this process.
Definition at line 3274 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_into_families()
| size_t gsc_split_into_families | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| size_t | maxentries_results, | ||
| gsc_GroupNum * | results | ||
| ) |
Split a group into families by their pedigrees.
Split a group into a set of smaller groups, each of which contains the genotypes in the original group that share a particular pair of parents. The number of new groups produced depends on the number of parent-combinations in the set of genotypes in the provided group.
Individuals with both parents unknown will be grouped together.
If more than maxentries_results groups are created by this function, only that many results will be saved into the results vector, though all the one-member groups will be created.
Stops executing if group is empty or has only one member.
Has short name: split_into_families
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split maxentries_results maximum number of group numbers that can be saved into the results vector. results Pointer to a vector into which to save the identifiers of the newly created family groups. Should have at least enough space for [maxentries_results] identifiers.
- Returns
- the number of families identified/number of family groups created. Also serves as the length of the array in results
Definition at line 2960 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_into_halfsib_families()
| size_t gsc_split_into_halfsib_families | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| const int | parent, | ||
| size_t | maxentries_results, | ||
| gsc_GroupNum * | results | ||
| ) |
Split a group into families of half-siblings by shared first or second parent.
Split a group into a set of smaller groups, each containing the genotypes from the original group that share one parent. The shared parent can be either the first or second parent, based on the value of the parameter parent. That is, if parent is 1, within the halfsib families produced, all genotypes will share the same first parent, but may have different second parents. The number of new groups produced depends on the number of unique first/second parents in the set of genotypes in the provided group.
Individuals with unknown parent will be grouped together.
Stops executing if group is empty or has only one member.
If more than maxentries_results groups are created by this function, only that many results will be saved into the results vector, though all the one-member groups will be created.
Has short name: split_into_halfsib_families
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split parent 1 to group together genotypes that share the same first parent, 2 group those with the same second parent. Raises an error if this parameter is not either of those values. maxentries_results maximum number of group numbers that can be saved into the results vector. results Pointer to a vector into which to save the identifiers of the newly created family groups. Should have at least enough space for [maxentries_results] identifiers.
- Returns
- the number of halfsib families identified/number of groups created.
Definition at line 2878 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_into_individuals()
| size_t gsc_split_into_individuals | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| size_t | maxentries_results, | ||
| gsc_GroupNum * | results | ||
| ) |
Split a group into n one-member groups.
Give every individual in the group a new group number that does not belong to any other existing group (thereby allocating each genotype in the group to a new group of 1).
Stops executing if group is empty or has only one member.
If more than maxentries_results groups are created by this function, only that many results will be saved into the results vector, though all the one-member groups will be created.
Has short name: split_into_individuals
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split maxentries_results maximum number of group numbers that can be saved into the results vector. results Pointer to a vector into which to save the identifiers of the newly created family groups. Should have at least enough space for [maxentries_results] identifiers.
- Returns
- the number of groups created (in this case, the same as the size of the original group). Also serves as the length of the array in results
Definition at line 3017 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_randomly_into_n()
| size_t gsc_split_randomly_into_n | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id, | ||
| const size_t | n, | ||
| gsc_GroupNum * | results | ||
| ) |
Allocate each member of the group to one of n groups with equal probability.
There is no guarantee that all groups will have members. There is no guarantee the groups will be near the same size.
Each genotype has equal probability of being allocated to each of n groups. The old group number (group_id) is included as one of these n possible groups.
To split by uneven probabilities instead: gsc_split_by_probabilities()
Has short name: split_randomly_into_n
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split n the number of groups among which to randomly distribute group members. results NULL if the caller does not care to know the identifiers of the groups created, or a pointer to an array to which these identifiers should be saved. It is assumed that the array is long enough to store n identifiers.
- Returns
- the number of groups created by this process.
Definition at line 3398 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_split_randomly_into_two()
| gsc_GroupNum gsc_split_randomly_into_two | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group_id | ||
| ) |
Flip a coin for each member of the group to decide if it should be moved to the new group.
There is no guarantee that there will be any genotypes in the new group (if all coin flips were 0) or any genotypes in the old group (if all coin flips were 1). There is no guarantee the two groups will be near the same size.
This could be useful for allocating a sex to genotypes.
A more general approach to this task: gsc_split_randomly_into_n()
An alternate approach to splitting a group in two: gsc_split_evenly_into_two()
Has short name: split_randomly_into_two
- Parameters
-
d the gsc_SimData struct on which to perform the operation group_id the group number of the group to be split
- Returns
- the group number of the new group to which some members of the old group may have been randomly allocated.
Definition at line 3333 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: