For iterating through the genotypes in the simulation. More...
 Collaboration diagram for Genotype Iterators:
Collaboration diagram for Genotype Iterators:Data Structures | |
| struct | gsc_GenoLocation |
| An gsc_AlleleMatrix/gsc_AlleleMatrix index coordinate of a particular genotype in the simulation. More... | |
| struct | gsc_ParentChoice |
| struct | gsc_BidirectionalIterator |
| A structure to iterate forwards and backwards through all genotypes in a gsc_SimData or through only the members of a group. More... | |
| struct | gsc_GappyIterator |
| A structure to iterate forwards through all positions in the gsc_AlleleMatrix linked list in gsc_SimData. More... | |
| struct | gsc_EmptyListNavigator |
| A structure to hold an initially empty AlleleMatrix list whose genotypes can be accessed sequentially by storage order. More... | |
| struct | gsc_RandomAccessIterator |
| A structure to search and cache indexes of all genotypes in a gsc_SimData or of all the members of a group. More... | |
Macros | |
| #define | GSC_INVALID_GENO_LOCATION (gsc_GenoLocation){.localAM=0,.localPos=GSC_NA_LOCALX} |
| Constant representing a nonexistent location in the simulation. More... | |
| #define | GSC_IS_VALID_LOCATION(g) (g.localAM != 0 && g.localPos != GSC_NA_LOCALX) |
| Check if a GenoLocation is INVALID_GENO_LOCATION. More... | |
Detailed Description
For iterating through the genotypes in the simulation.
It is possible to iterate through one group or through every genotype in the simulation.
Macro Definition Documentation
◆ GSC_INVALID_GENO_LOCATION
| #define GSC_INVALID_GENO_LOCATION (gsc_GenoLocation){.localAM=0,.localPos=GSC_NA_LOCALX} |
Constant representing a nonexistent location in the simulation.
Has short name: INVALID_GENO_LOCATION
Definition at line 1220 of file sim-operations.h.
◆ GSC_IS_VALID_LOCATION
| #define GSC_IS_VALID_LOCATION | ( | g | ) | (g.localAM != 0 && g.localPos != GSC_NA_LOCALX) |
Check if a GenoLocation is INVALID_GENO_LOCATION.
Has short name: IS_VALID_LOCATION
Definition at line 1224 of file sim-operations.h.
Function Documentation
◆ gsc_create_bidirectional_iter()
| gsc_BidirectionalIterator gsc_create_bidirectional_iter | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group | ||
| ) |
Create a bidirectional iterator.
A bidirectional iterator can be used to loop through members of a particular group or all genotypes in the simulation, forwards or backwards. The iterator is not initialised to any location at this point. The first call to a next* function will initialise it, or you can manually initialise using gsc_set_bidirectional_iter_to_start or gsc_set_bidirectional_iter_to_end. gsc_next_forwards will initialise it to the first in the group, or call gsc_next_backwards to initialise it to the last in the group.
Has short name: create_bidirectional_iter
- Warning
- An initialised iterator is only valid if no genotypes have been added to the group and no genotypes in the simulation as a whole have been removed since its creation. Discard the old iterator and create a new one when the state of the simulation changes.
- Parameters
-
d pointer to the gsc_SimData containing the genotypes to iterate through group the group number of the group to iterate through, or 0 to iterate through all genotypes.
- Returns
- uninitialised gsc_BidirectionalIterator for the provided group.
Definition at line 1444 of file sim-operations.c.
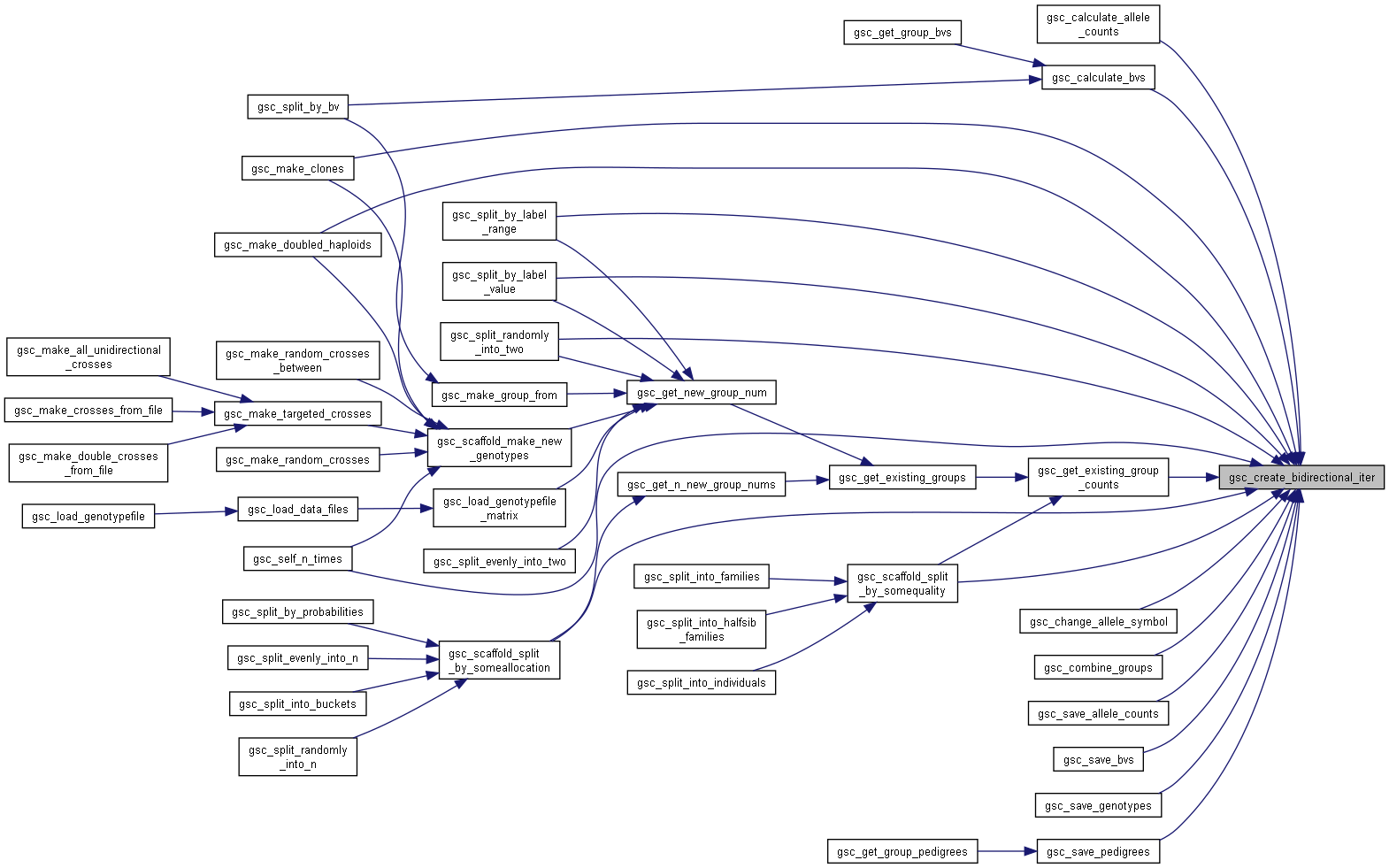
 Here is the call graph for this function:
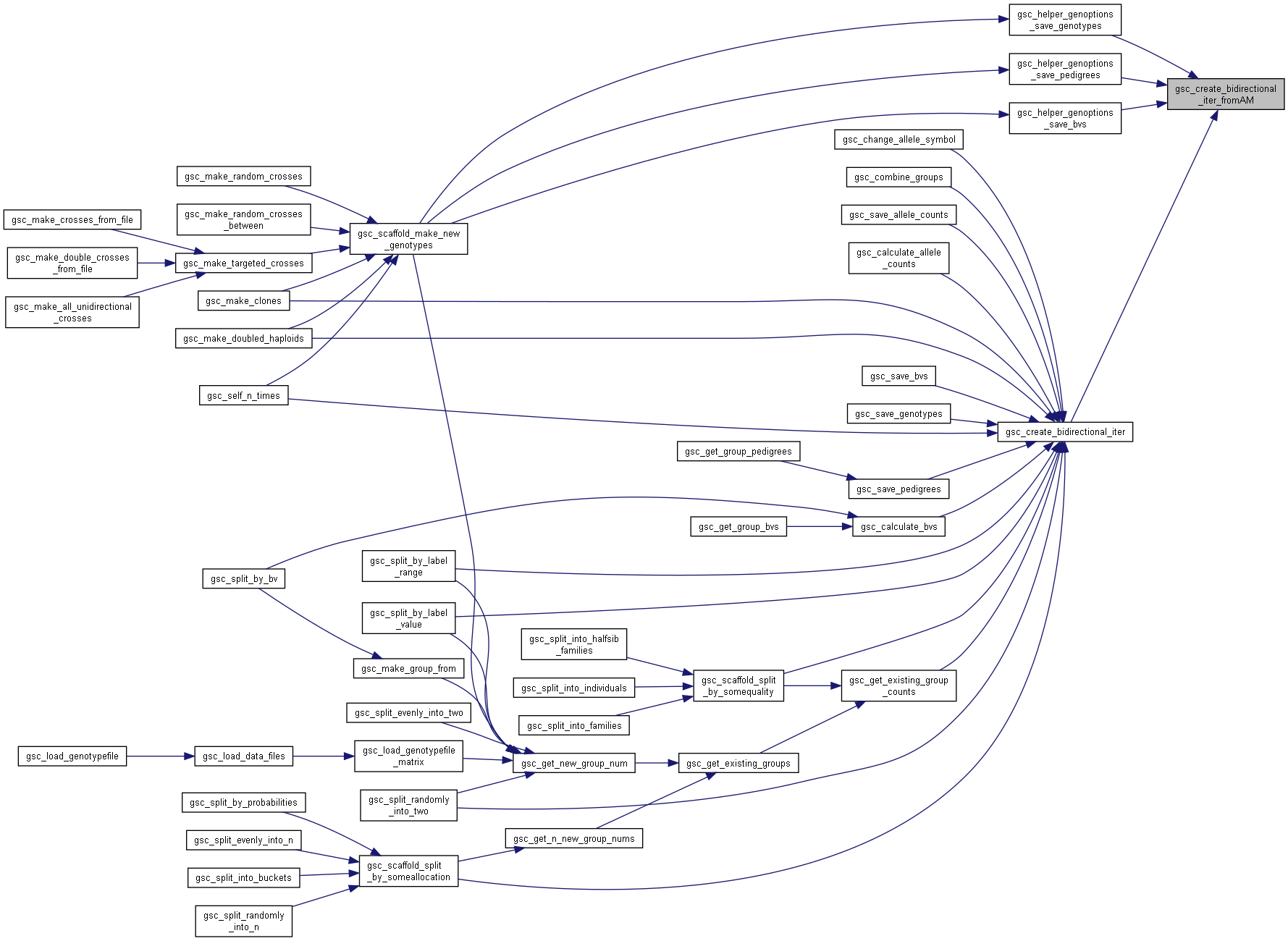
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_create_bidirectional_iter_fromAM()
| gsc_BidirectionalIterator gsc_create_bidirectional_iter_fromAM | ( | gsc_AlleleMatrix * | am, |
| const gsc_GroupNum | group | ||
| ) |
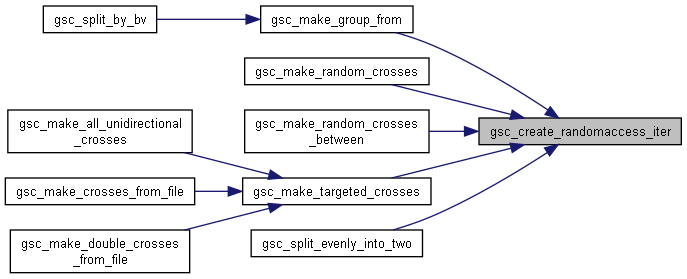
◆ gsc_create_randomaccess_iter()
| gsc_RandomAccessIterator gsc_create_randomaccess_iter | ( | gsc_SimData * | d, |
| const gsc_GroupNum | group | ||
| ) |
Create a Random Access Iterator.
A random access iterator and the function gsc_next_get_nth() can be used to get any genotype in the simulation by index or any genotype in a group by within-group index.
The random access iterator gives you access to the (i)th group member in O(n) time (n being the number of genotypes in the simulation), but contains a persistent cache that allows subsequent O(1) access to group members.
The random access iterator created by this function is initialised. If the groupSize attribute of the returned iterator is set to 0, no members of that group could be found in the simulation, so the iterator will never return a valid position.
Has short name: create_randomaccess_iter
- Warning
- An initialised iterator is only valid if no genotypes have been added to the group and no genotypes in the simulation as a whole have been removed since its creation. Discard the old iterator and create a new one when the state of the simulation changes.
- Parameters
-
d pointer to the gsc_SimData containing the genotypes to iterate through group the group number of the group to iterate through, or 0 to iterate through all genotypes.
- Returns
- initialised Random Access iterator for the provided group.
Definition at line 1491 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
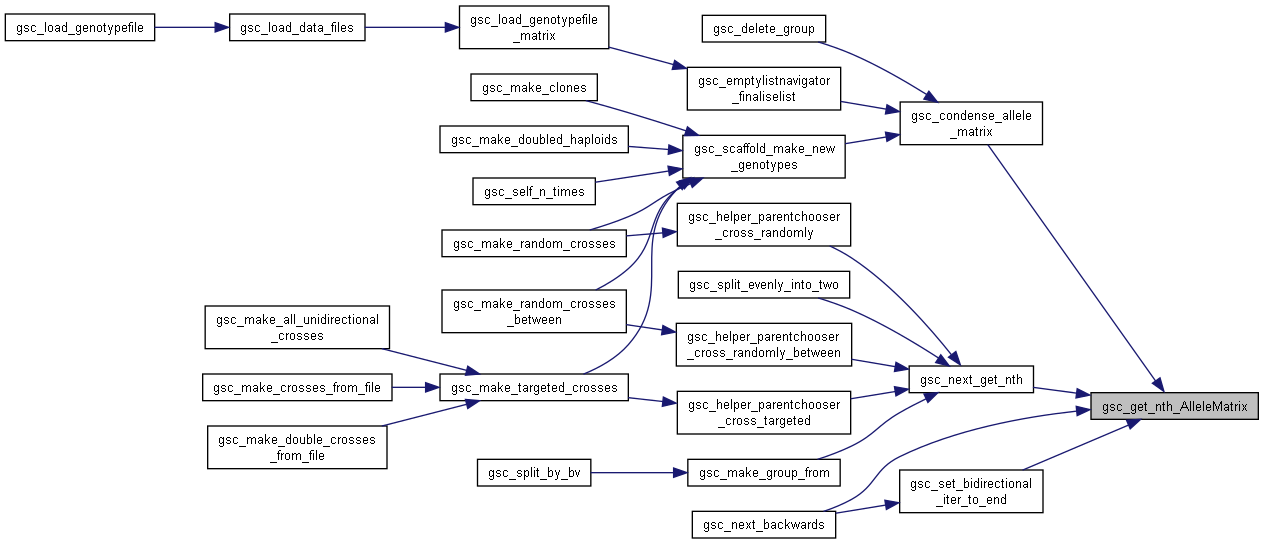
Here is the caller graph for this function:◆ gsc_get_nth_AlleleMatrix()
| gsc_AlleleMatrix * gsc_get_nth_AlleleMatrix | ( | gsc_AlleleMatrix * | listStart, |
| const unsigned int | n | ||
| ) |
Get an gsc_AlleleMatrix by index in the linked list.
listStart is considered the 0th gsc_AlleleMatrix (0-based indexing)
- Parameters
-
listStart the 0th gsc_AlleleMatrix in the linked list n the index of the desired gsc_AlleleMatrix in the linked list
- Returns
- pointer to the nth gsc_AlleleMatrix, if it exists in the list, or NULL if the list is shorter than n
Definition at line 1569 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_isValidLocation()
|
inlinestatic |
Identify whether a gsc_GenoLocation is INVALID_GENO_LOCATION.
- See also
- IS_VALID_LOCATION IS_VALID_LOCATION has the same function.
- Parameters
-
g location to check.
- Returns
- FALSE if g has either of the attributes of INVALID_GENO_LOCATION, TRUE otherwise
Definition at line 1233 of file sim-operations.h.
◆ gsc_next_backwards()
| gsc_GenoLocation gsc_next_backwards | ( | gsc_BidirectionalIterator * | it | ) |
Get the previous location from a bidirectional iterator.
Moves the pointer of a gsc_BidirectionalIterator backwards by one step, and returns the new location it points to. If the gsc_BidirectionalIterator is not initialised, then initialises it to the very last element.
Slightly slower than gsc_next_forwards, because the gsc_AlleleMatrix linked list is not bidirectional. To find the preceding gsc_AlleleMatrix, it needs to count forwards from the beginning of the list to find the n-1th gsc_AlleleMatrix.
Returns GSC_INVALID_GENO_LOCATION if the gsc_BidirectionalIterator is corrupted or if it is at the beginning of the sequence. Test the return value of this function with GSC_IS_VALID_LOCATION().
Has short name: next_backwards
- Parameters
-
it the gsc_BidirectionalIterator to iterate backwards
- Returns
- the location of the previous genotype in the sequence, or GSC_INVALID_GENO_LOCATION if the iterator is corrupted or the iterator's pointer is already at the first element.
Definition at line 1876 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_next_forwards()
| gsc_GenoLocation gsc_next_forwards | ( | gsc_BidirectionalIterator * | it | ) |
Get the next location from a bidirectional iterator.
Moves the pointer of a gsc_BidirectionalIterator forwards by one step, and returns the new location it points to. If the gsc_BidirectionalIterator is not initialised, then initialises it to the very first element.
Returns GSC_INVALID_GENO_LOCATION if the gsc_BidirectionalIterator is corrupted or if it is at the end of the sequence. Test the return value of this function with GSC_IS_VALID_LOCATION().
Has short name: next_forwards
- Parameters
-
it the gsc_BidirectionalIterator to iterate forwards
- Returns
- the location of the next genotype in the sequence, or GSC_INVALID_GENO_LOCATION if the iterator is corrupted or the iterator's pointer is already at the last element.
Definition at line 1762 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
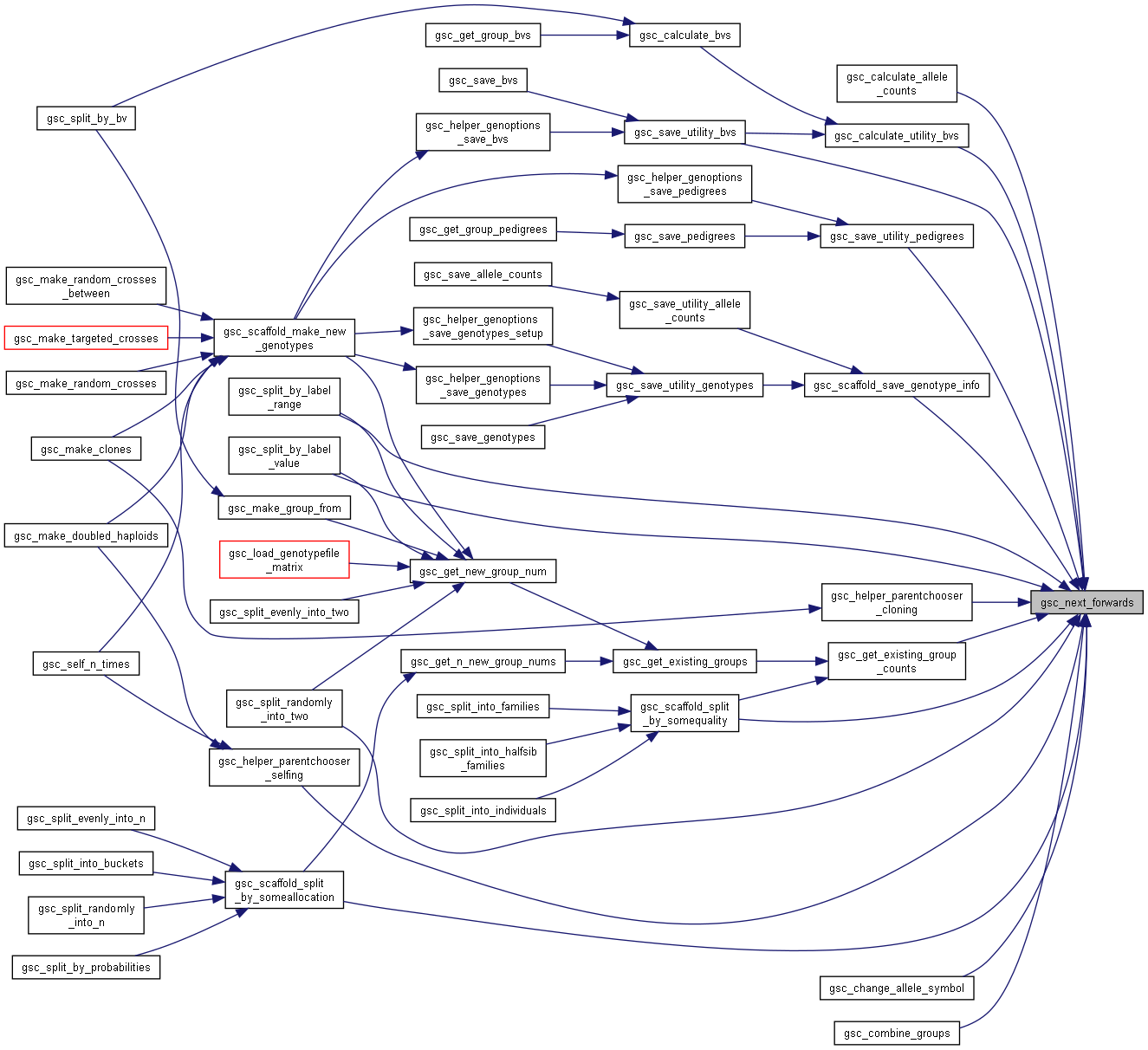
Here is the caller graph for this function:◆ gsc_next_get_nth()
| gsc_GenoLocation gsc_next_get_nth | ( | gsc_RandomAccessIterator * | it, |
| const unsigned int | n | ||
| ) |
Get a location by index using a gsc_RandomAccessIterator.
Gives the location of the provided global index (if it->group == 0) or the location of the provided group index (if it->group is not 0), by first searching the gsc_RandomAccessIterator's cache for it and if not, searching the gsc_SimData for it and adding it and its predecessors to the cache.
Returns GSC_INVALID_GENO_LOCATION if the iterator is corrupted or the index is invalid. Check the return value with GSC_IS_VALID_LOCATION().
Has short name: next_get_nth
- Parameters
-
it the gsc_RandomAccessIterator to read and update the cache of. n If the iterator is iterating through all genotypes, the global index in the simulation of the genotype you want to access. If the iterator is iterating through the group, the within-group index of the genotype you want to access. In either case the first genotype is at index 0.
- Returns
- the location of the nth genotype/nth group member, or GSC_INVALID_GENO_LOCATION if the index is invalid.
Definition at line 1997 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_set_bidirectional_iter_to_end()
| gsc_GenoLocation gsc_set_bidirectional_iter_to_end | ( | gsc_BidirectionalIterator * | it | ) |
Initialise a Bidirectional iterator to the end of its sequence.
Can be used to reset a gsc_BidirectionalIterator so that it is pointing at the very last member of the group it is looping through.
Has short name: set_bidirectional_iter_to_end
- Parameters
-
it BidirectioanlIterator to initialise
- Returns
- location of the last member of the sequence.
Definition at line 1671 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
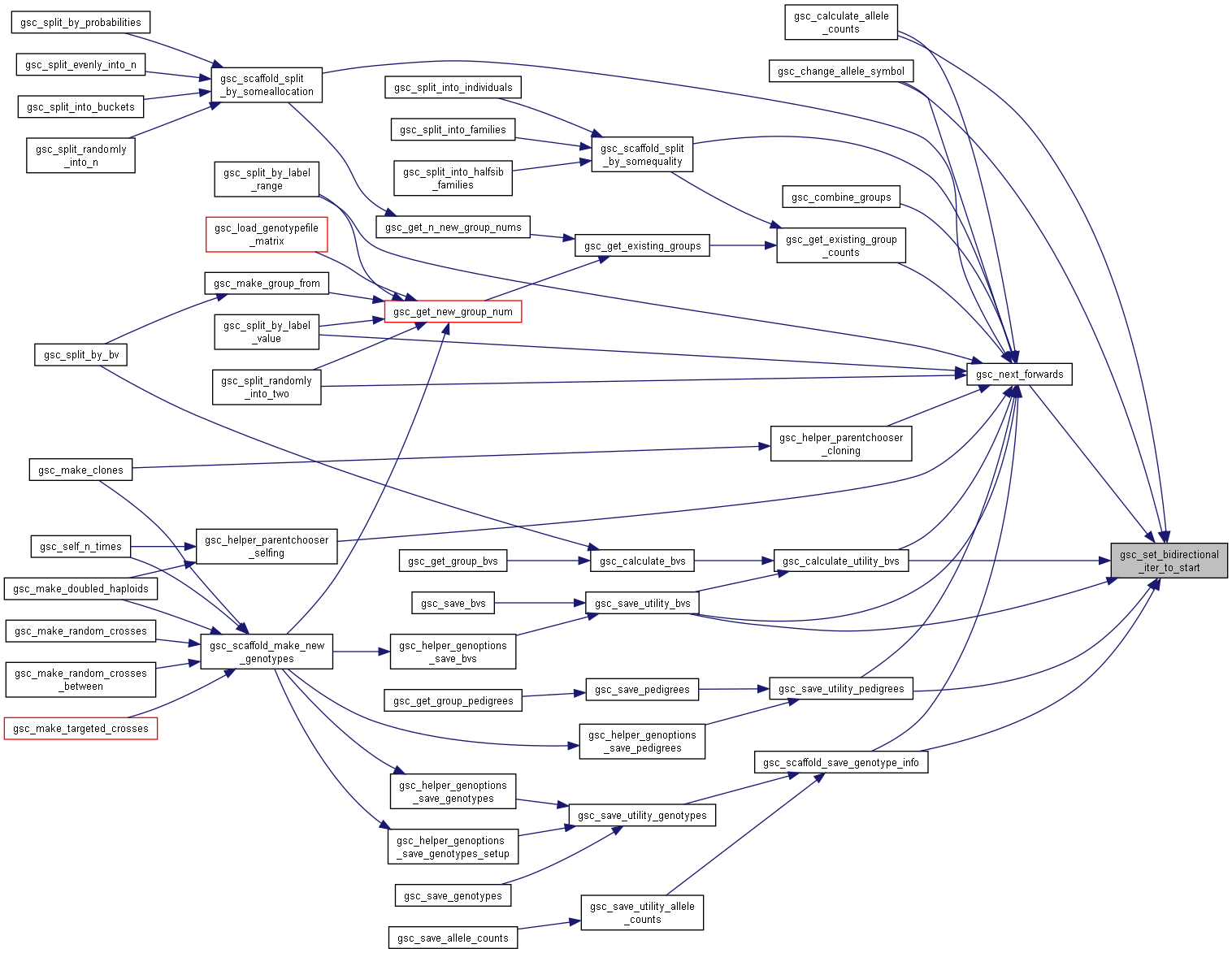
Here is the caller graph for this function:◆ gsc_set_bidirectional_iter_to_start()
| gsc_GenoLocation gsc_set_bidirectional_iter_to_start | ( | gsc_BidirectionalIterator * | it | ) |
Initialise a Bidirectional iterator to the start of its sequence.
Can be used to reset a gsc_BidirectionalIterator so that it is pointing at the very first member of the group it is looping through.
Has short name: set_bidirectional_iter_to_start
- Parameters
-
it BidirectioanlIterator to initialise
- Returns
- location of the first member of the sequence.
Definition at line 1593 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function: