For setup of the simulation (loading founders, genetic maps, and optionally allele effects). More...
Functions | |
| gsc_AlleleMatrix * | gsc_create_empty_allelematrix (const unsigned int n_markers, const unsigned int n_labels, const int *labelDefaults, const unsigned int n_genotypes) |
| Creator for an empty gsc_AlleleMatrix object of a given size. More... | |
| gsc_SimData * | gsc_create_empty_simdata (unsigned int RNGseed) |
| Creator for an empty gsc_SimData object on the heap. More... | |
| void | gsc_clear_simdata (gsc_SimData *d) |
| Clear a gsc_SimData object on the heap. More... | |
| gsc_GroupNum | gsc_load_genotypefile (gsc_SimData *d, const char *filename, const gsc_FileFormatSpec format) |
| Load a set of genotypes to a gsc_SimData object. More... | |
| gsc_MapID | gsc_load_mapfile (gsc_SimData *d, const char *filename) |
| Load a genetic map to a gsc_SimData object. More... | |
| gsc_MapID | gsc_create_recombmap_from_markerlist (gsc_SimData *d, unsigned int n_markers, struct gsc_MapfileUnit *markerlist) |
| Parse a list of markers/chrs/positions into a gsc_RecombinationMap and save to SimData. More... | |
| gsc_MapID | gsc_create_uniformspaced_recombmap (gsc_SimData *d, unsigned int n_markers, char **markernames, double expected_n_recombinations) |
| Create a uniformly-spaced gsc_RecombinationMap from a list of marker names and save to SimData. More... | |
| gsc_EffectID | gsc_load_effectfile (gsc_SimData *d, const char *filename) |
| Populates a gsc_SimData combination with effect values. More... | |
| struct gsc_MultiIDSet | gsc_load_data_files (gsc_SimData *d, const char *data_file, const char *map_file, const char *effect_file, const gsc_FileFormatSpec format) |
| Populates a gsc_SimData object with marker allele data, a genetic map, and (optionally) marker effect values. More... | |
Detailed Description
For setup of the simulation (loading founders, genetic maps, and optionally allele effects).
Function Documentation
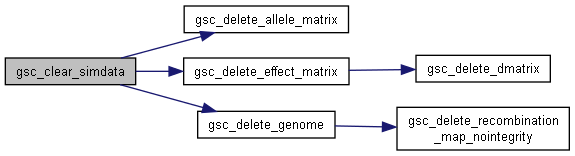
◆ gsc_clear_simdata()
| void gsc_clear_simdata | ( | gsc_SimData * | d | ) |
Clear a gsc_SimData object on the heap.
Has the effects of gsc_delete_simdata followed by gsc_create_empty_simdata, but guarantees the use of the same memory location for the new gsc_SimData.
Has short name: clear_simdata
- Parameters
-
d pointer to the gsc_SimData to be cleared.
Definition at line 142 of file sim-operations.c.
 Here is the call graph for this function:
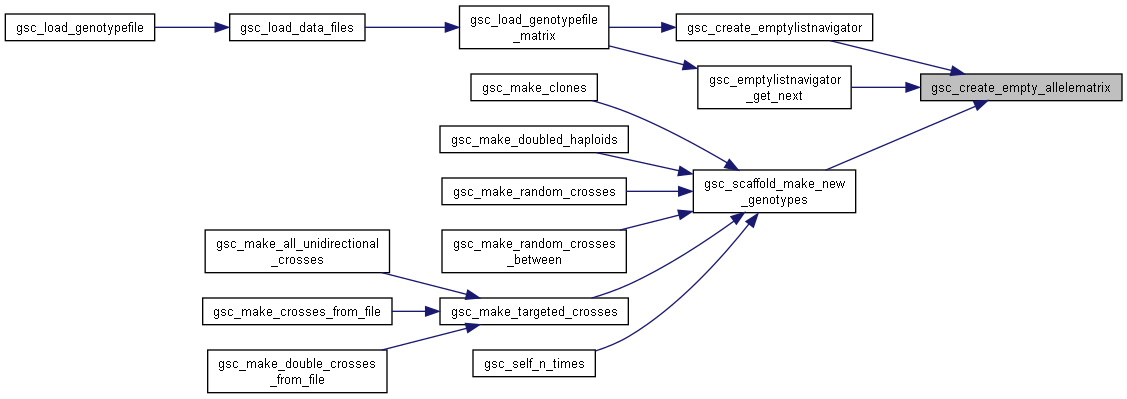
Here is the call graph for this function:◆ gsc_create_empty_allelematrix()
| gsc_AlleleMatrix * gsc_create_empty_allelematrix | ( | const unsigned int | n_markers, |
| const unsigned int | n_labels, | ||
| const int * | labelDefaults, | ||
| const unsigned int | n_genotypes | ||
| ) |
Creator for an empty gsc_AlleleMatrix object of a given size.
Includes memory allocation for n_genotypes worth of .alleles.
- Parameters
-
n_markers number of rows/markers to create n_labels number of custom labels to create labelDefaults an array of length [n_labels] of the default value to pre-fill for each custom label. Can be null if n_labels == 0. n_genotypes number of individuals to create. This includes filling the first n_genotypes entries of .alleles with heap char* of length n_markers, so that the alleles for these can be added without further memory allocation.
- Returns
- pointer to the empty created gsc_AlleleMatrix
Definition at line 62 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_create_empty_simdata()
| gsc_SimData * gsc_create_empty_simdata | ( | unsigned int | RNGseed | ) |
Creator for an empty gsc_SimData object on the heap.
This is the main struct that will contain/manage simulation data.
Has short name: create_empty_simdata
- Returns
- pointer to the empty created gsc_SimData
Definition at line 112 of file sim-operations.c.
 Here is the call graph for this function:
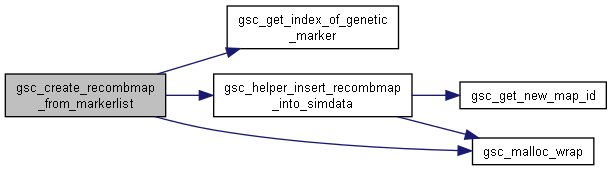
Here is the call graph for this function:◆ gsc_create_recombmap_from_markerlist()
| gsc_MapID gsc_create_recombmap_from_markerlist | ( | gsc_SimData * | d, |
| unsigned int | n_markers, | ||
| struct gsc_MapfileUnit * | markerlist | ||
| ) |
Parse a list of markers/chrs/positions into a gsc_RecombinationMap and save to SimData.
It partitions the provided list by chr, then sorts by position. Positions are interpreted as positions in cM for the purpose of calculating recombination probabilities between adjacent markers in the sorted list. Markers not present in the SimData's list of known markers are discarded. Markers with no names are discarded.
- Parameters
-
d SimData into which to load this RecombinationMap n_markers length of markerlist markerlist
- See also
- gsc_helper_parse_mapfile
- Parameters
-
n_chr If known, number of unique
- Returns
- gsc_MapID of the gsc_RecombinationMap that was just loaded into the simulation.
Definition at line 5641 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_create_uniformspaced_recombmap()
| gsc_MapID gsc_create_uniformspaced_recombmap | ( | gsc_SimData * | d, |
| unsigned int | n_markers, | ||
| char ** | markernames, | ||
| double | expected_n_recombinations | ||
| ) |
Create a uniformly-spaced gsc_RecombinationMap from a list of marker names and save to SimData.
The recombination map produced has one chromosome/linkage group. The markers in this chromosome are equally spaced and their ordering matches the ordering of markernames.
If markernames is NULL, n_markers is ignored and the uniformly-spaced recombination map is created from all markers listed in d->genome.
- Parameters
-
d SimData into which to load this RecombinationMap n_markers length of markernames markernames names of the markers to create this simple recombination map from. expected_n_recombinations
- Returns
- gsc_MapID of the gsc_RecombinationMap that was just loaded into the simulation.
Definition at line 5779 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_load_data_files()
| struct gsc_MultiIDSet gsc_load_data_files | ( | gsc_SimData * | d, |
| const char * | genotype_file, | ||
| const char * | map_file, | ||
| const char * | effect_file, | ||
| const gsc_FileFormatSpec | format | ||
| ) |
Populates a gsc_SimData object with marker allele data, a genetic map, and (optionally) marker effect values.
This is the suggested first function to call to set up a genomicSimulation simulation.
It will attempt to use file extensions to determine how to parse the files
Has short name: load_data_files
- Parameters
-
d pointer to gsc_SimData to be populated data_file string containing name/path of file containing SNP marker allele data. map_file string name/path of file containing genetic map data (optional, set to NULL if not needed). effect_file string name/path of file containing effect values (optional, set to NULL if not wanted). format details on the format of the file (option, set to GSC_DETECT_FILE_FORMAT to auto-detect format).
- Returns
- a gsc_MultiIDSet entry, containing the group number of the founding group, the map id of the recombination map loaded, and the effect set id of the loaded effect file if applicable.
Definition at line 7067 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_load_effectfile()
| gsc_EffectID gsc_load_effectfile | ( | gsc_SimData * | d, |
| const char * | filename | ||
| ) |
Populates a gsc_SimData combination with effect values.
If the gsc_SimData does not have a list of tracked markers already (i.e. no map data loaded), this function will be unable to match any marker names so will fail to load any marker effects.
The file's format should be:
marker allele eff
[marker] [allele] [effect]
[marker] [allele] [effect]
...
The header line is optional. If no header line is provided, it is assumed that the columns are in the above order (marker, then allele, then marker effect). If a header is provided, the columns "marker", "allele", and "eff" can be in any order.
Extra columns are not permitted. Columns may be space-separated or tab-separated. Column separators need not be consistent throughout the file, and may be made of multiple characters. That is, any consecutive sequence of spaces, tabs, and commas is interpreted as one column separator.
Marker names can be of any length and any characters excluding column separator (tab, space, or comma) and newline characters. Rows where the marker name does not match any marker in the SimData's tracked markers will be ignored.
Has short name: load_effectfile
- Parameters
-
d pointer to gsc_SimData to be populated. filename string name/path of file containing effect values.
- Returns
- the ID of the set of marker effects just loaded
Definition at line 5995 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_load_genotypefile()
| gsc_GroupNum gsc_load_genotypefile | ( | gsc_SimData * | d, |
| const char * | filename, | ||
| const gsc_FileFormatSpec | format | ||
| ) |
Load a set of genotypes to a gsc_SimData object.
Can be called on an empty SimData, in which case it invents a genetic map comprising of the markers present in the genotype file, all on a single chromosome and spaced 1cM apart, or on a SimData with existing maps or genotypes, in which case markers whose names do not appear in the SimData's list of tracked markers are discarded.
- See also
- enum gsc_GenotypeFileType for the types of files that can be loaded
Has short name: load_genotypefile
- Parameters
-
d pointer to gsc_SimData to be populated filename string name/path of file containing genotype data. format details on the format of the file (option, set to GSC_DETECT_FILE_FORMAT to auto-detect format).
Definition at line 7043 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_load_mapfile()
| gsc_MapID gsc_load_mapfile | ( | gsc_SimData * | d, |
| const char * | filename | ||
| ) |
Load a genetic map to a gsc_SimData object.
Can be called on an empty SimData, in which case it loads the first genetic map, sets up the list of known markers in the genome, and warns that no genotypes are loaded yet so many simulation functions will not yet be able to run. Can also be called on a SimData with existing map and genotypes, in which case the map file's contents are loaded as another recombination map.
The file's format should be:
marker chr pos
[marker name] [chr] [pos]
[marker name] [chr] [pos]
...
The header line is optional. If no header line is provided, it is assumed that the columns are in the above order (marker, then chromosome, then position). If a header is provided, the columns "marker", "chr", and "pos" can be in any order.
Map positions are assumped to be in centimorgans.
Extra columns are not permitted. Columns may be space-separated or tab-separated. Column separators need not be consistent throughout the file, and may be made of multiple characters. That is, any consecutive sequence of spaces, tabs, and commas is interpreted as one column separator.
Marker names can be of any length and any characters excluding column separator (tab, space, or comma) and newline characters.
Has short name: load_mapfile
- Parameters
-
d pointer to gsc_SimData to be populated filename string name/path of file containing genetic map data.
Definition at line 5906 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function: