Functions | |
| struct gsc_TableSize | gsc_get_file_dimensions (const char *filename, const char sep) |
Opens a table file and reads the number of columns and rows (including headers) separated by sep into a gsc_TableSize struct that is returned. More... | |
| unsigned int | gsc_get_from_ordered_pedigree_list (const gsc_PedigreeID target, const unsigned int listLen, const gsc_PedigreeID *list) |
| Binary search through list of unsigned integers. More... | |
| size_t | gsc_get_from_unordered_str_list (const char *target, const size_t listLen, const char **list) |
| Linear search through a list of strings. More... | |
| size_t | gsc_get_from_ordered_str_list (const char *target, const size_t listLen, const char **list) |
| Binary search through a list of strings. More... | |
| void | gsc_shuffle_up_to (rnd_pcg_t *rng, void *sequence, const size_t item_size, const size_t total_n, const size_t n_to_shuffle) |
| Produce a random ordering of the first n elements in an array using a (partial) Fisher-Yates shuffle. More... | |
| unsigned int | gsc_randomdraw_replacementrules (gsc_SimData *d, unsigned int max, unsigned int cap, unsigned int *member_uses, unsigned int noCollision) |
| Randomly pick a number in a range, optionally with a cap on how many times a number can be picked, and optionally required to be different to the last pick. More... | |
| gsc_LabelID | gsc_create_new_label (gsc_SimData *d, const int setTo) |
| Initialises a new custom label. More... | |
| void | gsc_change_label_default (gsc_SimData *d, const gsc_LabelID whichLabel, const int newDefault) |
| Set the default value of a custom label. More... | |
| void | gsc_change_label_to (gsc_SimData *d, const gsc_GroupNum whichGroup, const gsc_LabelID whichLabel, const int setTo) |
| Set the values of a custom label. More... | |
| void | gsc_change_label_by_amount (gsc_SimData *d, const gsc_GroupNum whichGroup, const gsc_LabelID whichLabel, const int byValue) |
| Increment the values of a custom label. More... | |
| void | gsc_change_label_to_values (gsc_SimData *d, const gsc_GroupNum whichGroup, const unsigned int startIndex, const gsc_LabelID whichLabel, const size_t n_values, const int *values) |
| Copy a vector of integers into a custom label. More... | |
| void | gsc_change_names_to_values (gsc_SimData *d, const gsc_GroupNum whichGroup, const unsigned int startIndex, const size_t n_values, const char **values) |
| Copy a vector of strings into the genotype name field. More... | |
| void | gsc_change_allele_symbol (gsc_SimData *d, const char *which_marker, const char from, const char to) |
| Replace all occurences of a given allele with a different symbol representation. More... | |
| int | gsc_get_integer_digits (const int i) |
Count and return the number of digits in i. More... | |
| unsigned int | gsc_get_index_of_label (const gsc_SimData *d, const gsc_LabelID label) |
| Function to identify the label lookup index of a label identifier. More... | |
| unsigned int | gsc_get_index_of_eff_set (const gsc_SimData *d, const gsc_EffectID eff_set_id) |
| Function to identify the lookup index of a marker effect set identifier. More... | |
| unsigned int | gsc_get_index_of_map (const gsc_SimData *d, const gsc_MapID map) |
| Function to identify the lookup index of a recombination map identifier. More... | |
| _Bool | gsc_get_index_of_genetic_marker (const char *target, gsc_KnownGenome g, unsigned int *out) |
| Return whether or not a marker name is present in the tracked markers, and at what index. More... | |
| gsc_LabelID | gsc_get_new_label_id (const gsc_SimData *d) |
| Function to identify the next sequential integer that is not already allocated to a label in the simulation. More... | |
| gsc_EffectID | gsc_get_new_eff_set_id (const gsc_SimData *d) |
| Function to identify the next sequential integer that is not already allocated to a marker effect set ID in the simulation. More... | |
| gsc_MapID | gsc_get_new_map_id (const gsc_SimData *d) |
| Function to identify the next sequential integer that is not already allocated to a map ID in the simulation. More... | |
| gsc_GroupNum | gsc_get_next_free_group_num (const size_t n_existing_groups, const gsc_GroupNum *existing_groups, size_t *cursor, gsc_GroupNum previous) |
| Iterator to get the next currently-free group number. More... | |
| gsc_GroupNum | gsc_get_new_group_num (gsc_SimData *d) |
| Function to identify the next sequential integer that does not identify a group that currently has member(s). More... | |
| void | gsc_get_n_new_group_nums (gsc_SimData *d, const size_t n, gsc_GroupNum *result) |
| Function to identify the next n sequential integers that do not identify a group that currently has member(s). More... | |
| void | gsc_condense_allele_matrix (gsc_SimData *d) |
| A function to tidy the internal storage of genotypes after addition or deletion of genotypes in the gsc_SimData. More... | |
Detailed Description
Function Documentation
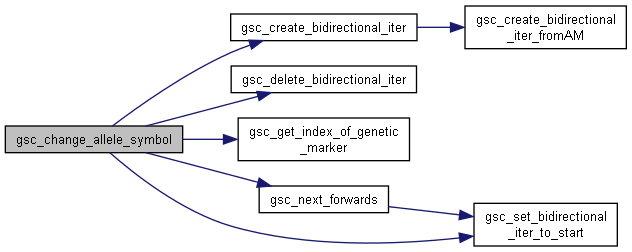
◆ gsc_change_allele_symbol()
| void gsc_change_allele_symbol | ( | gsc_SimData * | d, |
| const char * | which_marker, | ||
| const char | from, | ||
| const char | to | ||
| ) |
Replace all occurences of a given allele with a different symbol representation.
Alleles in genomicSimulation are represented by any single character. This function allows you to replace every instance of some allele with a different character. It may be used to replace unprintable alleles before saving output (like the null character '\0' used for a loaded founder genotype's alleles at a marker where no data was provided.)
If the allele is changed to a character which already represents another allele at that marker, the distinction between those two alleles will be lost.
If no marker name is provided, changes the allele symbol in all markers tracked by the simulation.
- Parameters
-
d SimData on which to perform the operation which_marker if null, any occurences of that allele symbol in any marker tracked by the simulation will be changed. If the name of a marker, replace all occurences of that allele symbol at that marker with the new symbol, and do not replace that allele symbol anywhere else. from character that currently represents the allele, whose representation is to be changed. to character to be the new representation of that allele
Definition at line 1041 of file sim-operations.c.
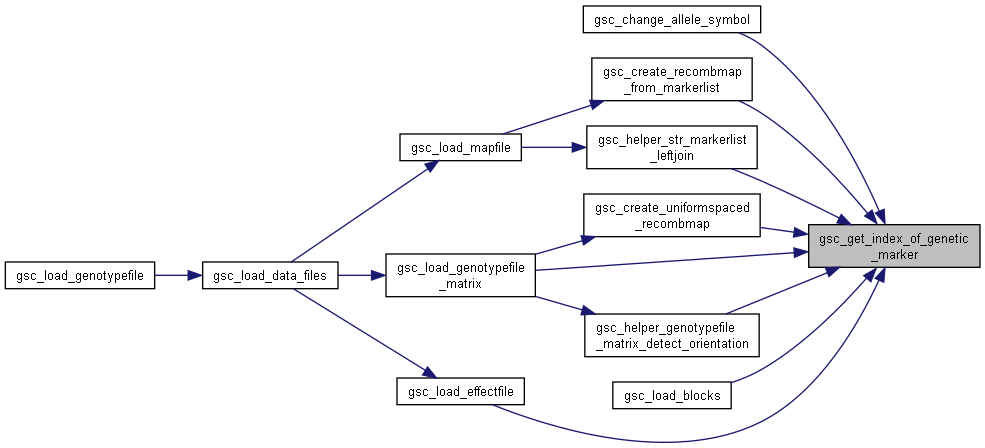
 Here is the call graph for this function:
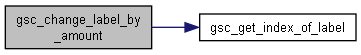
Here is the call graph for this function:◆ gsc_change_label_by_amount()
| void gsc_change_label_by_amount | ( | gsc_SimData * | d, |
| const gsc_GroupNum | whichGroup, | ||
| const gsc_LabelID | whichLabel, | ||
| const int | byValue | ||
| ) |
Increment the values of a custom label.
Increments the values of the custom label that has index whichLabel by the value byValue. Depending on the value of whichGroup, the function will modify the label of every single genotype in the simulation, or just modify the label of every member of a given group.
Has short name: change_label_by_amount
- Parameters
-
d pointer to the gsc_SimDatacontaining the genotypes and labels to be relabelledwhichGroup 0 to modify the relevant labels of all extant genotypes, or a positive integer to modify the relevant labels of all members of group `whichGroup. whichLabel the label id of the relevant label. byValue the value by which the appropriate labels will be incremented. For example, a value of 1 would increase all relevant labels by 1, a value of -2 would subtract 2 from each relevant label.
Definition at line 821 of file sim-operations.c.
 Here is the call graph for this function:
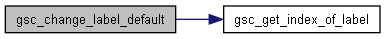
Here is the call graph for this function:◆ gsc_change_label_default()
| void gsc_change_label_default | ( | gsc_SimData * | d, |
| const gsc_LabelID | whichLabel, | ||
| const int | newDefault | ||
| ) |
Set the default value of a custom label.
Sets the default (birth) value of the custom label that has index whichLabel to the value newDefault.
Has short name: change_label_default
- Parameters
-
d pointer to the gsc_SimDatacontaining the genotypes and labels to be relabellewhichLabel the label id of the relevant label. newDefault the value to which the appropriate label's default will be set.
Definition at line 739 of file sim-operations.c.
 Here is the call graph for this function:
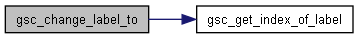
Here is the call graph for this function:◆ gsc_change_label_to()
| void gsc_change_label_to | ( | gsc_SimData * | d, |
| const gsc_GroupNum | whichGroup, | ||
| const gsc_LabelID | whichLabel, | ||
| const int | setTo | ||
| ) |
Set the values of a custom label.
Sets the values of the custom label that has index whichLabel to the value setTo. Depending on the value of whichGroup, the function will modify the label of every single genotype in the simulation, or just modify the label of every member of a given group.
Has short name: change_label_to
- Parameters
-
d pointer to the gsc_SimDatacontaining the genotypes and labels to be relabelledwhichGroup 0 to modify the relevant labels of all extant genotypes, or a positive integer to modify the relevant labels of all members of group whichGroup.whichLabel the label id of the relevant label. setTo the value to which the appropriate labels will be set.
Definition at line 765 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_change_label_to_values()
| void gsc_change_label_to_values | ( | gsc_SimData * | d, |
| const gsc_GroupNum | whichGroup, | ||
| const unsigned int | startIndex, | ||
| const gsc_LabelID | whichLabel, | ||
| const size_t | n_values, | ||
| const int * | values | ||
| ) |
Copy a vector of integers into a custom label.
Sets values of the custom label that has index whichLabel to the contents of the array values. The genotypes with the n_values contiguous simulation indexes starting with startIndex in group whichGroup are the ones given those labels. If whichGroup is 0, then it sets the labels of the genotypes with global indexes starting at startIndex.
If n_values is longer than the number of genotypes in the right group after startIndex, then the extra values are ignored.
Has short name: change_label_to_values
- Parameters
-
d pointer to the gsc_SimDatacontaining the genotypes and labels to be relabelledwhichGroup 0 to set the label of the genotypes with global indexes between startIndexandstartIndex + n_values, or a positive integer to set the label of thestartIndexth tostartIndex + n_valuesth members of groupwhichGroup.startIndex the first index of the group to set to a value whichLabel the label id of the relevant label. n_values length (number of entries) of the array valuesvalues vector of integers, of length at least [n_values], to paste into the chosen custom label of the chosen genotypes.
Definition at line 880 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_change_names_to_values()
| void gsc_change_names_to_values | ( | gsc_SimData * | d, |
| const gsc_GroupNum | whichGroup, | ||
| const unsigned int | startIndex, | ||
| const size_t | n_values, | ||
| const char ** | values | ||
| ) |
Copy a vector of strings into the genotype name field.
Sets genotype names to the contents of the array values. The genotypes with the n_values contiguous simulation indexes starting with startIndex in group whichGroup are the ones given those names. If whichGroup is 0, then it sets the names of the genotypes with global indexes starting at startIndex.
If n_values is longer than the number of genotypes in the right group after startIndex, then the extra values are ignored.
A deep copy of each name in values is made. This way, values can be a pointer to names of existing genotypes (say, if you are creating clones and want them to have the same names), and there will be no issues if only one of the genotypes sharing the name is deleted.
Has short name: change_names_to_values
- Parameters
-
d pointer to the gsc_SimDatacontaining the genotypes to be renamedwhichGroup 0 to set the names of the genotypes with global indexes between startIndexandstartIndex + n_values, or a positive integer to set the names of thestartIndexth tostartIndex + n_valuesth members of groupwhichGroup.startIndex the first index of the group to set to a value n_values length (number of entries) of valuesvalues vector of strings containing at least [n_values] strings, to paste into the name field of the chosen genotypes.
Definition at line 958 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function:◆ gsc_condense_allele_matrix()
| void gsc_condense_allele_matrix | ( | gsc_SimData * | d | ) |
A function to tidy the internal storage of genotypes after addition or deletion of genotypes in the gsc_SimData.
Not intended to be called by an end user - functions which require it should be calling it already.
Ideally, we want all gsc_AlleleMatrix structs in the gsc_SimData's linked list to have no gaps. That is, if there are more than CONTIG_WIDTH genotypes, the all gsc_AlleleMatrix structs except the last should be full (contain CONTIG_WIDTH genotypes), and the last should have the remaining n genotypes at local indexes 0 to n-1 (so all at the start of the gsc_AlleleMatrix, with no gaps between them).
This function will also clear any pre-allocated space that does not belong to any genotype (as determined by belonging to a index past AlleleMatrix->n_genotypes).
We trust that each gsc_AlleleMatrix's n_genotypes is correct.
This function achieves the cleanup by using two pointers: a checker out the front that identifies a genotype that needs to be shifted back/that occurs after a gap, and a filler that identifies each gap and copies the genotype at the checker back into it.
- Parameters
-
d The gsc_SimData struct on which to operate.
Definition at line 1360 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
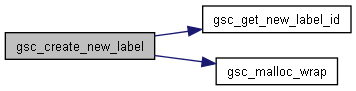
Here is the caller graph for this function:◆ gsc_create_new_label()
| gsc_LabelID gsc_create_new_label | ( | gsc_SimData * | d, |
| const int | setTo | ||
| ) |
Initialises a new custom label.
Creates a new custom label on every genotype currently and in future belonging to the gsc_SimData. The value of the label is set as setTo for every genotype.
Has short name: create_new_label
- Parameters
-
d pointer to the gsc_SimDatawhose childgsc_AlleleMatrixs will be given the new label.setTo the value to which every genotype's label is initialised.
- Returns
- the label id of the new label
Definition at line 622 of file sim-operations.c.
 Here is the call graph for this function:
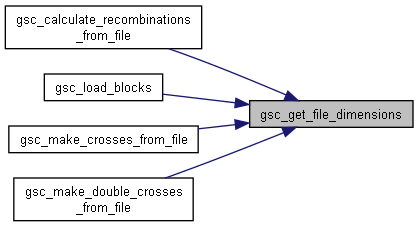
Here is the call graph for this function:◆ gsc_get_file_dimensions()
| struct gsc_TableSize gsc_get_file_dimensions | ( | const char * | filename, |
| const char | sep | ||
| ) |
Opens a table file and reads the number of columns and rows (including headers) separated by sep into a gsc_TableSize struct that is returned.
If the file fails to open, the simulation exits.
Rows must be either empty or have same number of columns as the first. Empty rows are not counted.
If a row with an invalid number of columns is found, the number of columns in the return value is set to 0. number of rows in the return value in this case is arbitrary.
- Parameters
-
filename the path/name to the table file whose dimensions we want sep the character that separates columns in the file eg tab
- Returns
- gsc_TableSize struct with .num_columns and .num_rows filled. These counts include header rows/columns and exclude blank rows.
Definition at line 234 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_from_ordered_pedigree_list()
| unsigned int gsc_get_from_ordered_pedigree_list | ( | const gsc_PedigreeID | target, |
| const unsigned int | listLen, | ||
| const gsc_PedigreeID * | list | ||
| ) |
Binary search through list of unsigned integers.
Returns the located index in an array of integers where the integer is target. Returns -1 if no match was found.
- See also
- gsc_get_from_unordered_str_list()
- gsc_get_from_ordered_pedigree_list()
- gsc_get_from_ordered_str_list()
The list is assumed to be sorted in ascending order. Only integers >0 are considered valid; entries of 0 are considered empty and can be located at any point in the list.
It uses a binary search method, but has to widen its search both directions if the desired midpoint has value 0.
- Parameters
-
target the integer to be located list an array of integers to search, with at least [list_len] entries list_len length of the array of integers to search
- Returns
- Index in
listwhere we find the same integer astarget, or -1 if no match is found. Binary search through a list of PedigreeIDs
Returns the located index in an array of gsc_PedigreeIDs where the gsc_PedigreeID is target. Returns GSC_NA_LOCALX if no match was found.
- See also
- gsc_get_from_unordered_str_list()
- gsc_get_from_ordered_uint_list()
- gsc_get_from_ordered_str_list()
The list is assumed to be sorted in ascending order. Only IDs >0 are considered valid; entries of 0 are considered empty and can be located at any point in the list.
It uses a binary search method, but has to widen its search both directions if the desired midpoint has value 0.
- Parameters
-
target the integer to be located list the array of integers to search, with at least [list_len] entries list_len length of the array of PedigreeIDs to search
- Returns
- Index in
listwhere we find the same integer astarget, or GSC_NA_LOCALX if no match is found.
Definition at line 390 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_from_ordered_str_list()
| size_t gsc_get_from_ordered_str_list | ( | const char * | target, |
| const size_t | listLen, | ||
| const char ** | list | ||
| ) |
Binary search through a list of strings.
Returns the first located index in an array of strings where the string is the same as the string target. Returns SIZE_MAX if no match was found.
- See also
- gsc_get_from_ordered_uint_list()
- gsc_get_from_ordered_pedigree_list()
- gsc_get_from_unordered_str_list()
The list of strings is assumed to be sorted in alphabetical order.
- Parameters
-
target the string to be located list the array of strings to search, with at least [list_len] entries list_len length of the array of strings to search
- Returns
- Index in
listwhere we find the same string astarget, or SIZE_MAX if no match is found.
Definition at line 490 of file sim-operations.c.
◆ gsc_get_from_unordered_str_list()
| size_t gsc_get_from_unordered_str_list | ( | const char * | target, |
| const size_t | listLen, | ||
| const char ** | list | ||
| ) |
Linear search through a list of strings.
Returns the first located index in an array of strings where the string is the same as the string target. Returns SIZE_MAX if no match was found.
- See also
- gsc_get_from_ordered_uint_list()
- gsc_get_from_ordered_pedigree_list()
- gsc_get_from_ordered_str_list()
The list of strings is not assumed to be sorted.
- Parameters
-
target the string to be located list the array of strings to search, with at least [list_len] entries list_len length of the array of strings to search
- Returns
- Index in
listwhere we find the same string astarget, or SIZE_MAX if no match is found.
Definition at line 463 of file sim-operations.c.
◆ gsc_get_index_of_eff_set()
| unsigned int gsc_get_index_of_eff_set | ( | const gsc_SimData * | d, |
| const gsc_EffectID | eff_set_id | ||
| ) |
Function to identify the lookup index of a marker effect set identifier.
- Parameters
-
d the gsc_SimData struct on which to perform the operation eff_set_id a marker effect set id
- Returns
- the index in d->e where the data for this effect set is stored, or GSC_NA_IDX if the effect set with that id could not be found.
Definition at line 3867 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_index_of_genetic_marker()
| _Bool gsc_get_index_of_genetic_marker | ( | const char * | target, |
| gsc_KnownGenome | g, | ||
| unsigned int * | out | ||
| ) |
Return whether or not a marker name is present in the tracked markers, and at what index.
- Parameters
-
target name of the marker that is to be located g genome containing list of tracked markers to search within out NULL if the output index is of no interest, or a pointer to a place to save a the index of the located marker in the KnownGenome object on success otherwise.
- Returns
- 1/truthy if the marker was located and its index saved to outindex, 0/falsy if the marker could not be located.
Definition at line 5201 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_index_of_label()
| unsigned int gsc_get_index_of_label | ( | const gsc_SimData * | d, |
| const gsc_LabelID | label | ||
| ) |
Function to identify the label lookup index of a label identifier.
- Parameters
-
d the gsc_SimData struct on which to perform the operation label a label id
- Returns
- the index in d->label_ids, d->label_defaults, and the ->labels table in gsc_AlleleMatrix where the data for this label is stored, or GSC_NA_IDX if the label with that id could not be found.
Definition at line 3835 of file sim-operations.c.
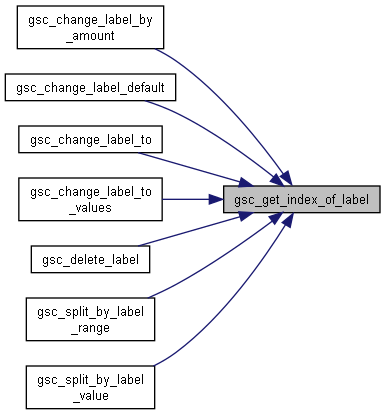
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_index_of_map()
| unsigned int gsc_get_index_of_map | ( | const gsc_SimData * | d, |
| const gsc_MapID | map | ||
| ) |
Function to identify the lookup index of a recombination map identifier.
- Parameters
-
d the simulation containing the map map a map id
- Returns
- the index in g->maps where the information for this map is stored, or GSC_NA_IDX if the map with that id could not be found.
Definition at line 3899 of file sim-operations.c.
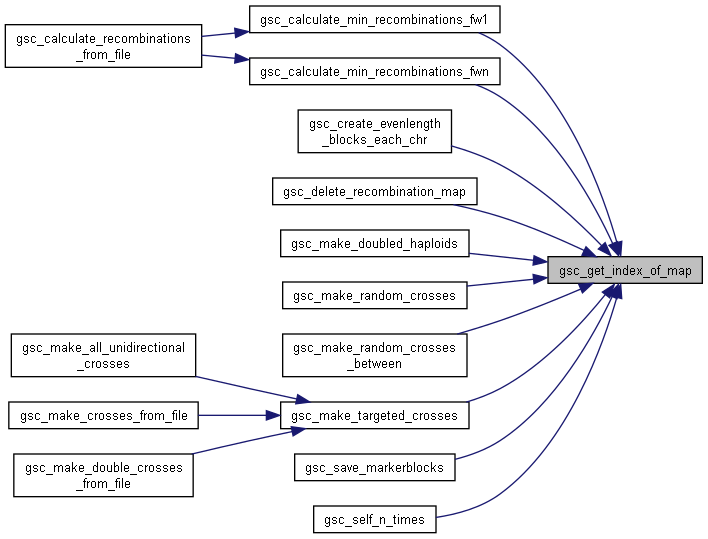
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_integer_digits()
| int gsc_get_integer_digits | ( | const int | i | ) |
Count and return the number of digits in i.
- Parameters
-
i the integer whose digits are to be counted.
- Returns
- the number of digits to print
i
Definition at line 1115 of file sim-operations.c.
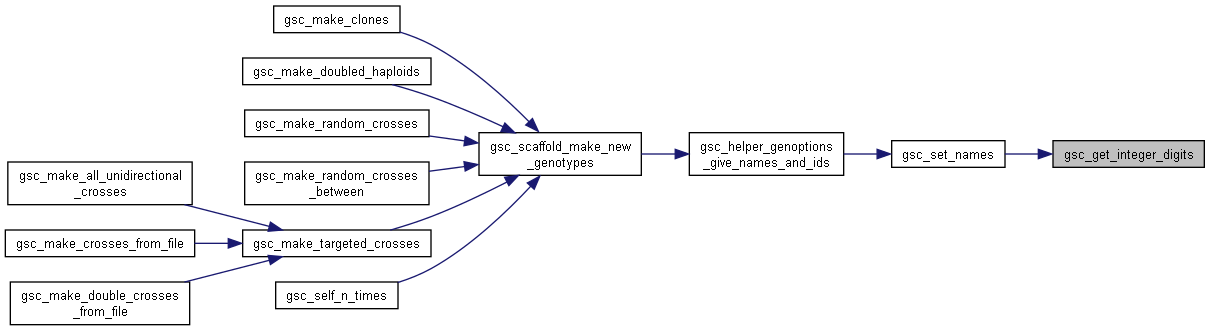
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_n_new_group_nums()
| void gsc_get_n_new_group_nums | ( | gsc_SimData * | d, |
| const size_t | n, | ||
| gsc_GroupNum * | result | ||
| ) |
Function to identify the next n sequential integers that do not identify a group that currently has member(s).
- Parameters
-
d the gsc_SimData struct on which to perform the operation n the number of group numbers to generate result pointer to an array of length at least n where the new group numbers generated can be saved.
Definition at line 3724 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_new_eff_set_id()
| gsc_EffectID gsc_get_new_eff_set_id | ( | const gsc_SimData * | d | ) |
Function to identify the next sequential integer that is not already allocated to a marker effect set ID in the simulation.
- Parameters
-
d the gsc_SimData struct on which to perform the operation
- Returns
- the next sequential currently-unused marker effect set id, an integer greater than 0.
Definition at line 3785 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_new_group_num()
| gsc_GroupNum gsc_get_new_group_num | ( | gsc_SimData * | d | ) |
Function to identify the next sequential integer that does not identify a group that currently has member(s).
This calls gsc_get_existing_groups() every time, so for better speed, functions that do repeated group creation, like gsc_split_into_individuals(), are recommended to use gsc_get_n_new_group_nums() (if they know the number of groups they need) or their own implementation, rather than calling this function repeatedly.
- Parameters
-
d the gsc_SimData struct on which to perform the operation
- Returns
- the next sequential currently-unused group number.
Definition at line 3690 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_new_label_id()
| gsc_LabelID gsc_get_new_label_id | ( | const gsc_SimData * | d | ) |
Function to identify the next sequential integer that is not already allocated to a label in the simulation.
- Parameters
-
d the gsc_SimData struct on which to perform the operation
- Returns
- the next sequential currently-unused label id, an integer greater than 0.
Definition at line 3761 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_new_map_id()
| gsc_MapID gsc_get_new_map_id | ( | const gsc_SimData * | d | ) |
Function to identify the next sequential integer that is not already allocated to a map ID in the simulation.
- Parameters
-
d the gsc_SimData struct on which to perform the operation
- Returns
- the next sequential currently-unused recombination map id, an integer greater than 0.
Definition at line 3809 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_get_next_free_group_num()
| gsc_GroupNum gsc_get_next_free_group_num | ( | const size_t | n_existing_groups, |
| const gsc_GroupNum * | existing_groups, | ||
| size_t * | cursor, | ||
| gsc_GroupNum | previous | ||
| ) |
Iterator to get the next currently-free group number.
In the vein of gsc_get_new_group_num or gsc_get_n_new_group_nums, a function that's effectively an iterator for unused group numbers. It's faster than calling either of those multiple times as it can re-use its 'cursor' position to not have to scan the array from the start every time, and it also gets to reuse a set of existing groups collected only once.
You will probably want to call gsc_get_existing_groups() before this one to get values for the parameters n_existing_groups and existing_groups. This is a function for internal use, so there won't be much/any bounds or error checking
- Parameters
-
n_existing_groups Length of the existing_groups array (eg, return value of gsc_get_existing_groups existing_groups Pointer to an array of gsc_GroupNums active in simulation (eg, output value of gsc_get_existing_groups). cursor Index in existing_groups that this function has currently checked up to. This value will be updated in the calling function. previous Last group number returned by this function, or GSC_NO_GROUP on first call.
- Returns
- the next sequential currently-unused (according to the memberships in existing_groups) group number.
Definition at line 3652 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_randomdraw_replacementrules()
| unsigned int gsc_randomdraw_replacementrules | ( | gsc_SimData * | d, |
| unsigned int | max, | ||
| unsigned int | cap, | ||
| unsigned int * | member_uses, | ||
| unsigned int | noCollision | ||
| ) |
Randomly pick a number in a range, optionally with a cap on how many times a number can be picked, and optionally required to be different to the last pick.
Used in random crossing functions.
Draws a random integer from the range [0, max). All numbers have equal probability of being drawn. The number will not be the same as noCollision (set noCollision to max or greater to make all numbers in the range possible results), and the number will fulfil member_uses[{number}] < cap, which can be used for selection without replacement or with only a certain number of replacements.
- Parameters
-
d gsc_SimData, only used as the source of the random number generator (in genomicSimulationC version). max upper bound (non-inclusive) of the range to draw from. cap maximum number of uses (as tracked by member_uses) of each number in the range. If, for a given number "num" in the range, member_uses["num"] is greater than or equal to cap, then the draw "num" will be discarded and the member_uses array of length max. See cap. noCollision this integer cannot be the return value.
- Returns
- Random integer from the range 0 (inclusive) to max (exclusive) that fulfils the cap and noCollision conditions, or GSC_NA_GLOBALX if input parameters made it impossible to draw any number.
Definition at line 8166 of file sim-operations.c.
 Here is the caller graph for this function:
Here is the caller graph for this function:◆ gsc_shuffle_up_to()
| void gsc_shuffle_up_to | ( | rnd_pcg_t * | rng, |
| void * | sequence, | ||
| const size_t | item_size, | ||
| const size_t | total_n, | ||
| const size_t | n_to_shuffle | ||
| ) |
Produce a random ordering of the first n elements in an array using a (partial) Fisher-Yates shuffle.
Modified from https://benpfaff.org/writings/clc/shuffle.html
After calling this function, the first 'n_to_shuffle' elements in the array will be randomly ordered by a Fischer-Yates shuffle. Every entry in the array could end up at any position, but the post-shuffle positions have only been calculated for the first 'n_to_shuffle' entries.
- Parameters
-
d gsc_SimData, only used for pointer to random number generator sequence the array item_size sizeof each element in the array total_n number of elements in the array n_to_shuffle the number of elements in the array to guarantee to be in randomly sorted order after the function is finished. (The remainder of the elements in the array will only be partially shuffled).
Definition at line 535 of file sim-operations.c.
 Here is the call graph for this function:
Here is the call graph for this function: Here is the caller graph for this function:
Here is the caller graph for this function: